Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002071A_C01 KMC002071A_c01
(542 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
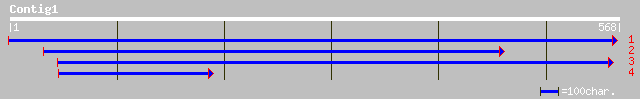
ref|NP_195890.1| putative protein; protein id: At5g02700.1 [Arab... 37 0.16
ref|NP_189482.1| hypothetical protein; protein id: At3g28410.1 [... 36 0.26
dbj|BAB02857.1| gb|AAF01596.1~gene_id:MFJ20.9~similar to unknown... 36 0.26
emb|CAA46367.1| putative DNA polymerase [Porphyra sp.] 33 1.7
gb|EAA22950.1| hypothetical protein [Plasmodium yoelii yoelii] 32 3.8
>ref|NP_195890.1| putative protein; protein id: At5g02700.1 [Arabidopsis thaliana]
gi|11357903|pir||T48291 hypothetical protein F9G14.10 -
Arabidopsis thaliana gi|7413545|emb|CAB86024.1| putative
protein [Arabidopsis thaliana]
Length = 456
Score = 37.0 bits (84), Expect = 0.16
Identities = 37/115 (32%), Positives = 56/115 (48%), Gaps = 6/115 (5%)
Frame = -3
Query: 510 NISSIEHVNFDI------HWKCEKYSFILLSWLVDLTNIKSLTVSTSTLQVLISIPHLLE 349
++SS+ N I + Y + L L N+K LTV + LQ+L S+ L
Sbjct: 255 DVSSLSEANLTIISSLLSPLTADGYQTMALEMLSKFHNVKRLTVGETLLQIL-SLAELRG 313
Query: 348 YKYPILGSLKSLKVKLRGDKYLFSSIPDGTVDYLLQNSPSAKVDIIRCST*YMHY 184
+P L +++L VK +++ S IP + LLQNSP K +R ST MH+
Sbjct: 314 VPFPTL-KVQTLTVK---TEFVRSVIPG--ISRLLQNSPGLKK--LRPSTMKMHH 360
>ref|NP_189482.1| hypothetical protein; protein id: At3g28410.1 [Arabidopsis
thaliana]
Length = 465
Score = 36.2 bits (82), Expect = 0.26
Identities = 32/103 (31%), Positives = 51/103 (49%), Gaps = 7/103 (6%)
Frame = -3
Query: 510 NISSIEHVNFDI-------HWKCEKYSFILLSWLVDLTNIKSLTVSTSTLQVLISIPHLL 352
++SS+ N +I ++Y + L L N+K LTV + LQ+L S+ L
Sbjct: 256 DVSSLSEANLNIISDRLLSPLTADRYQTMALEMLSKFHNVKRLTVGETILQIL-SLAELR 314
Query: 351 EYKYPILGSLKSLKVKLRGDKYLFSSIPDGTVDYLLQNSPSAK 223
+P L +++L VK +++ S IP + LLQNSP K
Sbjct: 315 GVPFPTL-KVQTLTVK---TEFVRSVIPG--ISRLLQNSPGLK 351
>dbj|BAB02857.1| gb|AAF01596.1~gene_id:MFJ20.9~similar to unknown protein
[Arabidopsis thaliana]
Length = 507
Score = 36.2 bits (82), Expect = 0.26
Identities = 32/103 (31%), Positives = 51/103 (49%), Gaps = 7/103 (6%)
Frame = -3
Query: 510 NISSIEHVNFDI-------HWKCEKYSFILLSWLVDLTNIKSLTVSTSTLQVLISIPHLL 352
++SS+ N +I ++Y + L L N+K LTV + LQ+L S+ L
Sbjct: 298 DVSSLSEANLNIISDRLLSPLTADRYQTMALEMLSKFHNVKRLTVGETILQIL-SLAELR 356
Query: 351 EYKYPILGSLKSLKVKLRGDKYLFSSIPDGTVDYLLQNSPSAK 223
+P L +++L VK +++ S IP + LLQNSP K
Sbjct: 357 GVPFPTL-KVQTLTVK---TEFVRSVIPG--ISRLLQNSPGLK 393
>emb|CAA46367.1| putative DNA polymerase [Porphyra sp.]
Length = 402
Score = 33.5 bits (75), Expect = 1.7
Identities = 16/50 (32%), Positives = 24/50 (48%)
Frame = -3
Query: 492 HVNFDIHWKCEKYSFILLSWLVDLTNIKSLTVSTSTLQVLISIPHLLEYK 343
H F I WKC+ S + ++ +I TV L+ + I HL+ YK
Sbjct: 34 HKPFAIGWKCDSLSITRFEYTQNVKDIHDYTVLIIFLREMFKIKHLIPYK 83
>gb|EAA22950.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 929
Score = 32.3 bits (72), Expect = 3.8
Identities = 22/83 (26%), Positives = 41/83 (48%), Gaps = 17/83 (20%)
Frame = -3
Query: 498 IEHVNFDIH------WKCEKYSFILLSWLVDLTNIKSLTVSTSTLQVLISIPHL------ 355
IE+ N D++ +K KY+F+ L W V T+I +L + TL++L ++ +
Sbjct: 815 IENYNSDLNNINISFFKISKYNFLYLLWNVINTHINNLYLRIITLKLLTNLTFIYIGKIT 874
Query: 354 -----LEYKYPILGSLKSLKVKL 301
+ + P+L + +K KL
Sbjct: 875 DNSVCIHFSLPLLKLINKIKKKL 897
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 431,642,544
Number of Sequences: 1393205
Number of extensions: 8632843
Number of successful extensions: 20381
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 19764
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20375
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18750593680
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)