Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002036A_C01 KMC002036A_c01
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
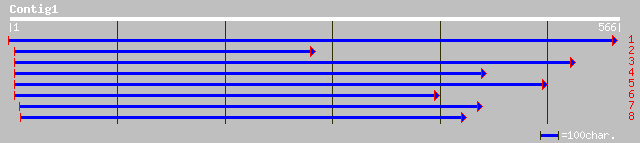
ref|NP_171893.1| G-box binding factor 4, GBF4; protein id: At1g0... 53 3e-06
gb|AAM63460.1| unknown [Arabidopsis thaliana] 49 3e-05
ref|NP_199221.1| bZIP transcription factor, G-box binding factor... 49 3e-05
pir||T04353 DNA binding protein - rice gi|1753085|gb|AAB39320.1|... 43 0.002
gb|AAF65459.1|AF245484_1 OSE2 [Oryza sativa] 43 0.002
>ref|NP_171893.1| G-box binding factor 4, GBF4; protein id: At1g03970.1, supported by
cDNA: 36980. [Arabidopsis thaliana]
gi|1169863|sp|P42777|GBF4_ARATH G-box binding factor 4
gi|25374029|pir||F86170 GBF4 [imported] - Arabidopsis
thaliana gi|403418|gb|AAA18414.1| GBF4
gi|4204292|gb|AAD10673.1| GBF4 [Arabidopsis thaliana]
gi|21593196|gb|AAM65145.1| G-box binding factor, GBF4
[Arabidopsis thaliana]
Length = 270
Score = 52.8 bits (125), Expect = 3e-06
Identities = 27/42 (64%), Positives = 33/42 (78%), Gaps = 1/42 (2%)
Frame = -1
Query: 557 LLKEKAERTKERFKQLMEEVIPVVEKRRPP-RPLRRINSMQW 435
LLKE E TKER+K+LME +IPV EK RPP RPL R +S++W
Sbjct: 229 LLKEIEESTKERYKKLMEVLIPVDEKPRPPSRPLSRSHSLEW 270
>gb|AAM63460.1| unknown [Arabidopsis thaliana]
Length = 315
Score = 49.3 bits (116), Expect = 3e-05
Identities = 25/43 (58%), Positives = 34/43 (78%), Gaps = 2/43 (4%)
Frame = -1
Query: 557 LLKEKAERTKERFKQLMEEVIPVVE--KRRPPRPLRRINSMQW 435
L KE ++ KER+++LME VIPVVE K++PPR LRRI S++W
Sbjct: 273 LSKEIEDKRKERYQKLMEFVIPVVEKPKQQPPRFLRRIRSLEW 315
>ref|NP_199221.1| bZIP transcription factor, G-box binding factor 4-related; protein
id: At5g44080.1, supported by cDNA: 25211., supported by
cDNA: gi_18175621, supported by cDNA: gi_20465650
[Arabidopsis thaliana] gi|9758567|dbj|BAB09068.1|
contains similarity to G-box binding
factor~gene_id:MRH10.19 [Arabidopsis thaliana]
gi|18175622|gb|AAL59898.1| unknown protein [Arabidopsis
thaliana] gi|20465651|gb|AAM20294.1| unknown protein
[Arabidopsis thaliana] gi|28317385|tpe|CAD29862.1| TPA:
basic leucine zipper transcription factor [Arabidopsis
thaliana]
Length = 315
Score = 49.3 bits (116), Expect = 3e-05
Identities = 25/43 (58%), Positives = 34/43 (78%), Gaps = 2/43 (4%)
Frame = -1
Query: 557 LLKEKAERTKERFKQLMEEVIPVVE--KRRPPRPLRRINSMQW 435
L KE ++ KER+++LME VIPVVE K++PPR LRRI S++W
Sbjct: 273 LSKEIEDKRKERYQKLMEFVIPVVEKPKQQPPRFLRRIRSLEW 315
>pir||T04353 DNA binding protein - rice gi|1753085|gb|AAB39320.1| leucine zipper
protein
Length = 217
Score = 43.1 bits (100), Expect = 0.002
Identities = 18/41 (43%), Positives = 29/41 (69%)
Frame = -1
Query: 557 LLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 435
+ KE+ E+ ++R K+L E V+PV+ ++ R LRR NSM+W
Sbjct: 177 MFKEQEEQHQKRLKELKEMVVPVIIRKTSARDLRRTNSMEW 217
>gb|AAF65459.1|AF245484_1 OSE2 [Oryza sativa]
Length = 217
Score = 43.1 bits (100), Expect = 0.002
Identities = 18/41 (43%), Positives = 29/41 (69%)
Frame = -1
Query: 557 LLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 435
+ KE+ E+ ++R K+L E V+PV+ ++ R LRR NSM+W
Sbjct: 177 MFKEQEEQHQKRLKELKEMVVPVIIRKTSARDLRRTNSMEW 217
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,020,151
Number of Sequences: 1393205
Number of extensions: 9286830
Number of successful extensions: 19789
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 19335
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19786
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)