Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002035A_C01 KMC002035A_c01
(602 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
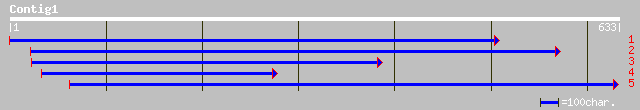
ref|NP_199397.1| putative protein; protein id: At5g45850.1 [Arab... 42 0.005
ref|NP_174218.1| unknown protein; protein id: At1g29240.1 [Arabi... 40 0.018
ref|NP_193598.1| putative protein; protein id: At4g18630.1 [Arab... 39 0.040
dbj|BAC42134.1| unknown protein [Arabidopsis thaliana] gi|289730... 39 0.040
ref|NP_180964.1| unknown protein; protein id: At2g34170.1 [Arabi... 39 0.068
>ref|NP_199397.1| putative protein; protein id: At5g45850.1 [Arabidopsis thaliana]
gi|9758932|dbj|BAB09313.1|
gene_id:K15I22.5~pir||T04855~similar to unknown protein
[Arabidopsis thaliana]
Length = 444
Score = 42.4 bits (98), Expect = 0.005
Identities = 18/32 (56%), Positives = 24/32 (74%)
Frame = -3
Query: 594 DSNSTSSNIKWETIVKTSKLNHDHVRYSQVTL 499
+ + S + KWETIVKTS +HDHVRYS+V +
Sbjct: 404 EKKTDSQSKKWETIVKTSYSHHDHVRYSEVCI 435
>ref|NP_174218.1| unknown protein; protein id: At1g29240.1 [Arabidopsis thaliana]
Length = 609
Score = 40.4 bits (93), Expect = 0.018
Identities = 27/65 (41%), Positives = 38/65 (57%), Gaps = 2/65 (3%)
Frame = -3
Query: 567 KWETIVKTSKLNHDHVRYSQVTLL-IPLAIILSSSTLVMTKMTINSNFLCRN*PC-INSS 394
KWETIVKTS ++ DH+RYS+V++ + LS + L + + + C C SS
Sbjct: 546 KWETIVKTSYMHRDHIRYSEVSISHFHIYSALSKNQLAIFGIYLLQ--FCAGTGCSYISS 603
Query: 393 IQNLR 379
IQNLR
Sbjct: 604 IQNLR 608
>ref|NP_193598.1| putative protein; protein id: At4g18630.1 [Arabidopsis thaliana]
gi|7486275|pir||T04855 hypothetical protein F28A21.40 -
Arabidopsis thaliana gi|4539382|emb|CAB37448.1| putative
protein [Arabidopsis thaliana]
gi|7268657|emb|CAB78865.1| putative protein [Arabidopsis
thaliana]
Length = 472
Score = 39.3 bits (90), Expect = 0.040
Identities = 16/25 (64%), Positives = 21/25 (84%)
Frame = -3
Query: 567 KWETIVKTSKLNHDHVRYSQVTLLI 493
KWETIVKTS +++DH RYSQV + +
Sbjct: 448 KWETIVKTSYVHNDHARYSQVLITL 472
>dbj|BAC42134.1| unknown protein [Arabidopsis thaliana] gi|28973001|gb|AAO63825.1|
unknown protein [Arabidopsis thaliana]
Length = 479
Score = 39.3 bits (90), Expect = 0.040
Identities = 16/26 (61%), Positives = 21/26 (80%)
Frame = -3
Query: 567 KWETIVKTSKLNHDHVRYSQVTLLIP 490
KWETIVKTS +++DH RYSQ ++ P
Sbjct: 448 KWETIVKTSYVHNDHARYSQELIVYP 473
>ref|NP_180964.1| unknown protein; protein id: At2g34170.1 [Arabidopsis thaliana]
gi|25408333|pir||C84753 hypothetical protein At2g34170
[imported] - Arabidopsis thaliana
gi|20196904|gb|AAM14830.1| unknown protein [Arabidopsis
thaliana]
Length = 523
Score = 38.5 bits (88), Expect = 0.068
Identities = 18/29 (62%), Positives = 22/29 (75%)
Frame = -3
Query: 594 DSNSTSSNIKWETIVKTSKLNHDHVRYSQ 508
+ NSTS KWETIVKTS + DH+RYS+
Sbjct: 484 EENSTSVT-KWETIVKTSYTHRDHIRYSE 511
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 465,760,314
Number of Sequences: 1393205
Number of extensions: 9131733
Number of successful extensions: 18890
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 18535
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18884
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)