Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002007A_C01 KMC002007A_c01
(551 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
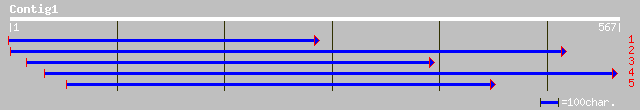
ref|NP_568917.1| putative protein; protein id: At5g59960.1, supp... 119 2e-26
gb|AAM61471.1| unknown [Arabidopsis thaliana] 119 2e-26
dbj|BAB03617.1| P0009G03.17 [Oryza sativa (japonica cultivar-gro... 117 1e-25
gb|AAL92603.1|AC115678_4 similar to Dictyostelium discoideum (Sl... 47 1e-04
dbj|BAB08364.1| gene_id:MMN10.21~unknown protein [Arabidopsis th... 37 0.16
>ref|NP_568917.1| putative protein; protein id: At5g59960.1, supported by cDNA:
120982., supported by cDNA: gi_13605840 [Arabidopsis
thaliana]
Length = 359
Score = 119 bits (298), Expect = 2e-26
Identities = 52/66 (78%), Positives = 58/66 (87%)
Frame = -2
Query: 550 RAVRHAAWDALDFLFPVGPYPRHLISLFFRLLYPWYWPSSCWYFAISCLQAIFNSLLRLI 371
RAVRHAAWDALD LFPVG YPRHLISLFFRLLYPWYWPSSCW F +SC++A+ S++RLI
Sbjct: 290 RAVRHAAWDALDSLFPVGRYPRHLISLFFRLLYPWYWPSSCWNFVVSCIKAVLYSIVRLI 349
Query: 370 FSSWEK 353
FS EK
Sbjct: 350 FSRREK 355
>gb|AAM61471.1| unknown [Arabidopsis thaliana]
Length = 359
Score = 119 bits (298), Expect = 2e-26
Identities = 52/66 (78%), Positives = 58/66 (87%)
Frame = -2
Query: 550 RAVRHAAWDALDFLFPVGPYPRHLISLFFRLLYPWYWPSSCWYFAISCLQAIFNSLLRLI 371
RAVRHAAWDALD LFPVG YPRHLISLFFRLLYPWYWPSSCW F +SC++A+ S++RLI
Sbjct: 290 RAVRHAAWDALDSLFPVGRYPRHLISLFFRLLYPWYWPSSCWNFVVSCIKAVLYSIVRLI 349
Query: 370 FSSWEK 353
FS EK
Sbjct: 350 FSRREK 355
>dbj|BAB03617.1| P0009G03.17 [Oryza sativa (japonica cultivar-group)]
Length = 230
Score = 117 bits (292), Expect = 1e-25
Identities = 48/73 (65%), Positives = 59/73 (80%)
Frame = -2
Query: 550 RAVRHAAWDALDFLFPVGPYPRHLISLFFRLLYPWYWPSSCWYFAISCLQAIFNSLLRLI 371
RAVRHAAW+ LD LFPVG YPRH+ISLFFRLLYPWYWPSSCW F ++C++ ++ +L LI
Sbjct: 153 RAVRHAAWNTLDLLFPVGRYPRHVISLFFRLLYPWYWPSSCWNFIMTCVKTVYYYILNLI 212
Query: 370 FSSWEKIAKPKSQ 332
SSWE + +P Q
Sbjct: 213 VSSWENMRRPNHQ 225
>gb|AAL92603.1|AC115678_4 similar to Dictyostelium discoideum (Slime mold). MkpA protein
Length = 837
Score = 47.4 bits (111), Expect = 1e-04
Identities = 22/51 (43%), Positives = 35/51 (68%)
Frame = -2
Query: 550 RAVRHAAWDALDFLFPVGPYPRHLISLFFRLLYPWYWPSSCWYFAISCLQA 398
R+VR + + LD LFP G + RH ++LFFRLL+P+Y S ++ IS +++
Sbjct: 781 RSVRTKSREVLDRLFPDGKFSRHTVNLFFRLLHPYYSIGSIIHWGISIIKS 831
>dbj|BAB08364.1| gene_id:MMN10.21~unknown protein [Arabidopsis thaliana]
Length = 290
Score = 37.0 bits (84), Expect = 0.16
Identities = 16/17 (94%), Positives = 16/17 (94%)
Frame = -2
Query: 550 RAVRHAAWDALDFLFPV 500
RAVRHAAWDALD LFPV
Sbjct: 270 RAVRHAAWDALDSLFPV 286
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 491,584,490
Number of Sequences: 1393205
Number of extensions: 10575389
Number of successful extensions: 24196
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 23495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24193
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)