Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002005A_C01 KMC002005A_c01
(770 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
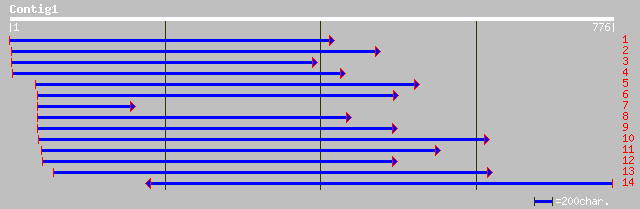
sp|Q01899|HS7M_PHAVU HEAT SHOCK 70 KD PROTEIN, MITOCHONDRIAL PRE... 312 4e-84
sp|P37900|HS7M_PEA HEAT SHOCK 70 KD PROTEIN, MITOCHONDRIAL PRECU... 306 3e-82
pir||S19140 dnaK-type molecular chaperone PHSP1 precursor, mitoc... 306 3e-82
gb|AAB26551.1| HSP68=68 kda heat-stress DnaK homolog [Lycopersic... 288 6e-77
ref|NP_196521.1| heat shock protein 70 (Hsc70-5); protein id: At... 285 7e-76
>sp|Q01899|HS7M_PHAVU HEAT SHOCK 70 KD PROTEIN, MITOCHONDRIAL PRECURSOR
gi|100004|pir||S25005 dnaK-type molecular chaperone
precursor, mitochondrial - kidney bean
gi|22636|emb|CAA47345.1| 70 kDa heat shock protein
[Phaseolus vulgaris]
Length = 675
Score = 312 bits (799), Expect = 4e-84
Identities = 162/176 (92%), Positives = 167/176 (94%)
Frame = -2
Query: 769 ELVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIEKMVK 590
+LVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLS+DEIEKMVK
Sbjct: 501 DLVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSEDEIEKMVK 560
Query: 589 EAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKEIEDAVSELRTAMS 410
EAELHAQKDQERK LIDIRNSADT+IYSIEKSLGEYR+KIPSE AKEIEDAVS+LR AMS
Sbjct: 561 EAELHAQKDQERKTLIDIRNSADTTIYSIEKSLGEYREKIPSETAKEIEDAVSDLRKAMS 620
Query: 409 GDNADEIKSKLDAANKAVSKIGEHMSGGGSSGGSSTGGGSQGGDQAPEAEYEEVKK 242
GDN DEIKSKLDAANKAVSKIGEHMS GGSSGGSS GG GGDQAPEAEYEEVKK
Sbjct: 621 GDNVDEIKSKLDAANKAVSKIGEHMS-GGSSGGSSAGGSQGGGDQAPEAEYEEVKK 675
>sp|P37900|HS7M_PEA HEAT SHOCK 70 KD PROTEIN, MITOCHONDRIAL PRECURSOR
gi|20835|emb|CAA38536.1| HSP70 [Pisum sativum]
Length = 675
Score = 306 bits (783), Expect = 3e-82
Identities = 157/176 (89%), Positives = 169/176 (95%)
Frame = -2
Query: 769 ELVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIEKMVK 590
+LVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEI+KMVK
Sbjct: 502 DLVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIDKMVK 561
Query: 589 EAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKEIEDAVSELRTAMS 410
EAELHAQ+DQERKALIDIRNSADTSIYSIEKSL EYR+KIP+EVAKEIEDAVS+LRTAM+
Sbjct: 562 EAELHAQRDQERKALIDIRNSADTSIYSIEKSLAEYREKIPAEVAKEIEDAVSDLRTAMA 621
Query: 409 GDNADEIKSKLDAANKAVSKIGEHMSGGGSSGGSSTGGGSQGGDQAPEAEYEEVKK 242
G+NAD+IK+KLDAANKAVSKIG+HMSGG S G S GGSQGG+QAPEAEYEEVKK
Sbjct: 622 GENADDIKAKLDAANKAVSKIGQHMSGGSSGGPSE--GGSQGGEQAPEAEYEEVKK 675
>pir||S19140 dnaK-type molecular chaperone PHSP1 precursor, mitochondrial - garden
pea
Length = 675
Score = 306 bits (783), Expect = 3e-82
Identities = 157/176 (89%), Positives = 169/176 (95%)
Frame = -2
Query: 769 ELVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIEKMVK 590
+LVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEI+KMVK
Sbjct: 502 DLVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIDKMVK 561
Query: 589 EAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKEIEDAVSELRTAMS 410
EAELHAQ+DQERKALIDIRNSADTSIYSIEKSL EYR+KIP+EVAKEIEDAVS+LRTAM+
Sbjct: 562 EAELHAQRDQERKALIDIRNSADTSIYSIEKSLAEYREKIPAEVAKEIEDAVSDLRTAMA 621
Query: 409 GDNADEIKSKLDAANKAVSKIGEHMSGGGSSGGSSTGGGSQGGDQAPEAEYEEVKK 242
G+NAD+IK+KLDAANKAVSKIG+HMSGG S G S GGSQGG+QAPEAEYEEVKK
Sbjct: 622 GENADDIKAKLDAANKAVSKIGQHMSGGSSGGPSE--GGSQGGEQAPEAEYEEVKK 675
>gb|AAB26551.1| HSP68=68 kda heat-stress DnaK homolog [Lycopersicon
peruvianum=tomatoes, Peptide Mitochondrial Partial, 580
aa]
Length = 580
Score = 288 bits (737), Expect = 6e-77
Identities = 145/176 (82%), Positives = 162/176 (91%)
Frame = -2
Query: 769 ELVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIEKMVK 590
ELVGIPPAPRG+PQIEVTFDIDANG+VTVSAKDK+T KEQQITIRSSGGLS+DEI+KMV+
Sbjct: 406 ELVGIPPAPRGMPQIEVTFDIDANGMVTVSAKDKATSKEQQITIRSSGGLSEDEIDKMVR 465
Query: 589 EAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKEIEDAVSELRTAMS 410
EAE+HAQKDQERKALIDIRNSADT+IYSIEKSL EY+DK+P EV EIE A+S+LR AM
Sbjct: 466 EAEMHAQKDQERKALIDIRNSADTTIYSIEKSLSEYKDKVPKEVVTEIETAISDLRAAMG 525
Query: 409 GDNADEIKSKLDAANKAVSKIGEHMSGGGSSGGSSTGGGSQGGDQAPEAEYEEVKK 242
+N D+IK KLDAANKAVSKIGEHM+ GGSSGG+S GGG+QGGDQ PEAEYEEVKK
Sbjct: 526 TENIDDIKPKLDAANKAVSKIGEHMA-GGSSGGASGGGGAQGGDQPPEAEYEEVKK 580
>ref|NP_196521.1| heat shock protein 70 (Hsc70-5); protein id: At5g09590.1, supported
by cDNA: gi_6746589 [Arabidopsis thaliana]
gi|11277110|pir||T49939 heat shock protein 70 (Hsc70-5) -
Arabidopsis thaliana gi|6746590|gb|AAF27638.1|AF217458_1
heat shock protein 70 [Arabidopsis thaliana]
gi|7671430|emb|CAB89371.1| heat shock protein 70
(Hsc70-5) [Arabidopsis thaliana]
Length = 682
Score = 285 bits (728), Expect = 7e-76
Identities = 144/177 (81%), Positives = 168/177 (94%), Gaps = 1/177 (0%)
Frame = -2
Query: 769 ELVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIEKMVK 590
+LVGIPP+PRG+PQIEVTFDIDANGIVTVSAKDK+TGK QQITIRSSGGLS+D+I+KMV+
Sbjct: 507 DLVGIPPSPRGVPQIEVTFDIDANGIVTVSAKDKTTGKVQQITIRSSGGLSEDDIQKMVR 566
Query: 589 EAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKEIEDAVSELRTAMS 410
EAELHAQKD+ERK LID +N+ADT+IYSIEKSLGEYR+KIPSE+AKEIEDAV++LR+A S
Sbjct: 567 EAELHAQKDKERKELIDTKNTADTTIYSIEKSLGEYREKIPSEIAKEIEDAVADLRSASS 626
Query: 409 GDNADEIKSKLDAANKAVSKIGEHMSGGGSSGGSSTGGGSQGG-DQAPEAEYEEVKK 242
GD+ +EIK+K++AANKAVSKIGEHMS GGS GGS+ GGGS+GG DQAPEAEYEEVKK
Sbjct: 627 GDDLNEIKAKIEAANKAVSKIGEHMS-GGSGGGSAPGGGSEGGSDQAPEAEYEEVKK 682
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 620,659,937
Number of Sequences: 1393205
Number of extensions: 13550131
Number of successful extensions: 138797
Number of sequences better than 10.0: 1636
Number of HSP's better than 10.0 without gapping: 67740
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 116516
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37815044670
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)