Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001990A_C01 KMC001990A_c01
(666 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
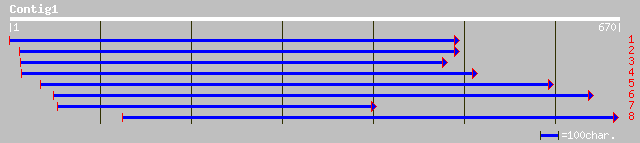
sp|P19954|RR30_SPIOL Plastid-specific 30S ribosomal protein 1, c... 159 3e-38
gb|AAA34039.1| ribosomal protein 30S subunit 159 3e-38
ref|NP_568447.1| putative protein; protein id: At5g24490.1, supp... 156 2e-37
gb|ZP_00107001.1| hypothetical protein [Nostoc punctiforme] 89 5e-17
ref|NP_487224.1| light-repressed protein [Nostoc sp. PCC 7120] g... 88 1e-16
>sp|P19954|RR30_SPIOL Plastid-specific 30S ribosomal protein 1, chloroplast precursor
(CS-S5) (CS5) (S22) (Ribosomal protein 1) (PSRP-1)
gi|279640|pir||R3SPS5 ribosomal protein CS-S22
precursor, chloroplast - spinach
gi|12316|emb|CAA41960.1| chloroplast ribosomal protein
S22 [Spinacia oleracea] gi|18031|emb|CAA33403.1| spinach
S22 r-protein [Spinacia oleracea]
Length = 302
Score = 159 bits (402), Expect = 3e-38
Identities = 81/129 (62%), Positives = 99/129 (75%), Gaps = 5/129 (3%)
Frame = -2
Query: 665 VSDHGRHMKGFNRAKVREPVEVEVPLEE-----DDDISPLEEEGEATLVEVVRTKYFDMP 501
VSDHGRHMKGFNR+KVR+P V + EE + +P+ E + + EVVRTKYFDMP
Sbjct: 175 VSDHGRHMKGFNRSKVRDPEPVRITREEVLEEVESAPAPVSVEDDDFIEEVVRTKYFDMP 234
Query: 500 PLTVSEAIEQLVNVDHAFYAFRNEETGEINIVYKRKEGGYGLIIPKGNGQAEKLEPLMVE 321
PLT++EA+EQL NVDH FYAFRNEETG+INI+YKRKEGGYGLIIPK +G+ EKLE L V+
Sbjct: 235 PLTITEAVEQLENVDHDFYAFRNEETGDINILYKRKEGGYGLIIPK-DGKTEKLESLPVQ 293
Query: 320 PAREASLKE 294
++ S E
Sbjct: 294 TDKQPSFAE 302
>gb|AAA34039.1| ribosomal protein 30S subunit
Length = 302
Score = 159 bits (402), Expect = 3e-38
Identities = 81/129 (62%), Positives = 99/129 (75%), Gaps = 5/129 (3%)
Frame = -2
Query: 665 VSDHGRHMKGFNRAKVREPVEVEVPLEE-----DDDISPLEEEGEATLVEVVRTKYFDMP 501
VSDHGRHMKGFNR+KVR+P V + EE + +P+ E + + EVVRTKYFDMP
Sbjct: 175 VSDHGRHMKGFNRSKVRDPEPVRITREEVLEEVESAPAPVSVEDDDFIEEVVRTKYFDMP 234
Query: 500 PLTVSEAIEQLVNVDHAFYAFRNEETGEINIVYKRKEGGYGLIIPKGNGQAEKLEPLMVE 321
PLT++EA+EQL NVDH FYAFRNEETG+INI+YKRKEGGYGLIIPK +G+ EKLE L V+
Sbjct: 235 PLTITEAVEQLENVDHDFYAFRNEETGDINILYKRKEGGYGLIIPK-DGKTEKLESLPVQ 293
Query: 320 PAREASLKE 294
++ S E
Sbjct: 294 TDKQPSFAE 302
>ref|NP_568447.1| putative protein; protein id: At5g24490.1, supported by cDNA:
gi_13877770 [Arabidopsis thaliana]
gi|13877771|gb|AAK43963.1|AF370148_1 unknown protein
[Arabidopsis thaliana]
Length = 308
Score = 156 bits (395), Expect = 2e-37
Identities = 84/126 (66%), Positives = 95/126 (74%), Gaps = 3/126 (2%)
Frame = -2
Query: 662 SDHGRHMKGFNRAKVREPV--EVEVPLEEDDDISPLEEEGEATLV-EVVRTKYFDMPPLT 492
SDHGRHMKGFNR KVREPV V +E+ D S EEE E L+ E+VRTK F+MPPLT
Sbjct: 183 SDHGRHMKGFNRLKVREPVIEPVVEDVEDSTDSSVGEEEEEDDLIKEIVRTKTFEMPPLT 242
Query: 491 VSEAIEQLVNVDHAFYAFRNEETGEINIVYKRKEGGYGLIIPKGNGQAEKLEPLMVEPAR 312
V+EA+EQL V H FY F+NEETGEINIVYKRKEGGYGLIIPK +G+AEK+EPL E
Sbjct: 243 VAEAVEQLELVSHDFYGFQNEETGEINIVYKRKEGGYGLIIPKKDGKAEKVEPLPTEQLN 302
Query: 311 EASLKE 294
E S E
Sbjct: 303 EHSFAE 308
>gb|ZP_00107001.1| hypothetical protein [Nostoc punctiforme]
Length = 214
Score = 89.0 bits (219), Expect = 5e-17
Identities = 41/61 (67%), Positives = 50/61 (81%), Gaps = 1/61 (1%)
Frame = -2
Query: 533 EVVRTKYFDMPPLTVSEAIEQLVNVDHAFYAFRNEETGEINIVYKRKEGGYGLIIPK-GN 357
EVVRTKYF MPP+T++EA+EQL V H FY FRN ETGEIN++Y+R GGYG+I P+ GN
Sbjct: 131 EVVRTKYFSMPPMTLAEALEQLQLVGHDFYMFRNSETGEINVIYERNHGGYGVIHPRNGN 190
Query: 356 G 354
G
Sbjct: 191 G 191
>ref|NP_487224.1| light-repressed protein [Nostoc sp. PCC 7120]
gi|25535366|pir||AI2203 light-repressed protein
[imported] - Nostoc sp. (strain PCC 7120)
gi|17132279|dbj|BAB74883.1| light-repressed protein
[Nostoc sp. PCC 7120]
Length = 213
Score = 87.8 bits (216), Expect = 1e-16
Identities = 39/59 (66%), Positives = 47/59 (79%)
Frame = -2
Query: 533 EVVRTKYFDMPPLTVSEAIEQLVNVDHAFYAFRNEETGEINIVYKRKEGGYGLIIPKGN 357
EVVRTKYF MPP+TV+EA+EQL V H FY F N ETGEIN++Y+R GGYG+I P+ N
Sbjct: 132 EVVRTKYFSMPPMTVTEALEQLQLVGHDFYMFHNAETGEINVIYERNHGGYGVIQPRNN 190
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 574,808,729
Number of Sequences: 1393205
Number of extensions: 13039536
Number of successful extensions: 69077
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 47829
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 60820
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)