Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001984A_C01 KMC001984A_c01
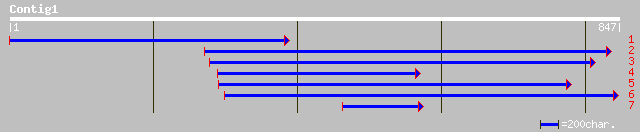
(819 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S59538 heat shock transcription factor HSF34 - soybean gi|6... 104 2e-21
pir||S71851 heat shock transcription factor HSF4 - Arabidopsis t... 42 0.011
ref|NP_195416.1| heat shock transcription factor HSF4; protein i... 42 0.011
ref|XP_045716.1| similar to KIAA1819 protein [Homo sapiens] 38 0.16
gb|AAK93833.1| MECT1/MAML2 fusion protein [Homo sapiens] 38 0.16
>pir||S59538 heat shock transcription factor HSF34 - soybean
gi|662930|emb|CAA87077.1| heat shock transcription
factor 34 [Glycine max]
Length = 282
Score = 104 bits (259), Expect = 2e-21
Identities = 59/99 (59%), Positives = 73/99 (73%), Gaps = 1/99 (1%)
Frame = -2
Query: 818 VGPDQIERIMRKGSYGSDENNAVGEGKCEGEGECLKLFGVWLKEKSFCEKRNN-KRERED 642
VGPDQI+RIMR+GS GS+ N VGEG G G+CLKLFGVWLK + +KRNN KR RED
Sbjct: 196 VGPDQIDRIMRQGSCGSE--NVVGEG---GGGDCLKLFGVWLKGDTLTDKRNNHKRGRED 250
Query: 641 QMGFGGPPIPAKVSKTTTVVDFGAVNVKAVMNSSSKVCS 525
QMGFGGP ++ ++ VVDFGAVN +M S++VC+
Sbjct: 251 QMGFGGP----RLKESKPVVDFGAVN---IMMKSNRVCN 282
>pir||S71851 heat shock transcription factor HSF4 - Arabidopsis thaliana
gi|1619921|gb|AAC31756.1| heat shock transcription
factor 4 [Arabidopsis thaliana]
Length = 284
Score = 42.0 bits (97), Expect = 0.011
Identities = 34/106 (32%), Positives = 53/106 (49%), Gaps = 8/106 (7%)
Frame = -2
Query: 818 VGPDQIERIMRKGSYG---SDENNAV-----GEGKCEGEGECLKLFGVWLKEKSFCEKRN 663
V P+QI+++++ G + SDE + G G EG GE LKLFGVWLK +
Sbjct: 194 VRPEQIDKMIKGGKFKPVESDEESECEGCDGGGGAEEGVGEGLKLFGVWLKGE------R 247
Query: 662 NKREREDQMGFGGPPIPAKVSKTTTVVDFGAVNVKAVMNSSSKVCS 525
KR+R+++ V + + + V+ A + SSKVC+
Sbjct: 248 KKRDRDEK---------NYVVSGSRMTEIKNVDFHAPLWKSSKVCN 284
>ref|NP_195416.1| heat shock transcription factor HSF4; protein id: At4g36990.1
[Arabidopsis thaliana] gi|12643794|sp|Q96320|HSF4_ARATH
Heat shock factor protein 4 (HSF 4) (Heat shock
transcription factor 4) (HSTF 4) gi|25296104|pir||H85436
heat shock transcription factor HSF4 [imported] -
Arabidopsis thaliana gi|2464881|emb|CAB16764.1| heat
shock transcription factor HSF4 [Arabidopsis thaliana]
gi|3256070|emb|CAA74398.1| Heat Shock Factor 4
[Arabidopsis thaliana] gi|7270648|emb|CAB80365.1| heat
shock transcription factor HSF4 [Arabidopsis thaliana]
gi|21539531|gb|AAM53318.1| heat shock transcription
factor HSF4 [Arabidopsis thaliana]
gi|28059096|gb|AAO30002.1| heat shock transcription
factor HSF4 [Arabidopsis thaliana]
Length = 284
Score = 42.0 bits (97), Expect = 0.011
Identities = 34/106 (32%), Positives = 53/106 (49%), Gaps = 8/106 (7%)
Frame = -2
Query: 818 VGPDQIERIMRKGSYG---SDENNAV-----GEGKCEGEGECLKLFGVWLKEKSFCEKRN 663
V P+QI+++++ G + SDE + G G EG GE LKLFGVWLK +
Sbjct: 194 VRPEQIDKMIKGGKFKPVESDEESECEGCDGGGGAEEGVGEGLKLFGVWLKGE------R 247
Query: 662 NKREREDQMGFGGPPIPAKVSKTTTVVDFGAVNVKAVMNSSSKVCS 525
KR+R+++ V + + + V+ A + SSKVC+
Sbjct: 248 KKRDRDEK---------NYVVSGSRMTEIKNVDFHAPLWKSSKVCN 284
>ref|XP_045716.1| similar to KIAA1819 protein [Homo sapiens]
Length = 928
Score = 38.1 bits (87), Expect = 0.16
Identities = 23/46 (50%), Positives = 27/46 (58%)
Frame = -2
Query: 518 KQNTLDRHTYSIHNQKIVFSGLFCLGKKEANKEIRGEAQSCSKHVE 381
K NTL HT+S N GLF +G KE KE GE SCSKH++
Sbjct: 14 KTNTLPSHTHSPGN------GLFNMGLKEVKKE-PGETLSCSKHMD 52
>gb|AAK93833.1| MECT1/MAML2 fusion protein [Homo sapiens]
Length = 1024
Score = 38.1 bits (87), Expect = 0.16
Identities = 23/46 (50%), Positives = 27/46 (58%)
Frame = -2
Query: 518 KQNTLDRHTYSIHNQKIVFSGLFCLGKKEANKEIRGEAQSCSKHVE 381
K NTL HT+S N GLF +G KE KE GE SCSKH++
Sbjct: 113 KTNTLPSHTHSPGN------GLFNMGLKEVKKE-PGETLSCSKHMD 151
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 722,849,992
Number of Sequences: 1393205
Number of extensions: 16375921
Number of successful extensions: 44712
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 41478
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44518
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42296827742
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)