Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001962A_C01 KMC001962A_c01
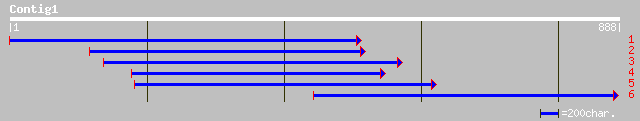
(883 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q43082|HEM3_PEA Porphobilinogen deaminase, chloroplast precur... 289 4e-77
gb|AAM64573.1| hydroxymethylbilane synthase [Arabidopsis thaliana] 270 2e-71
ref|NP_196445.1| hydroxymethylbilane synthase; protein id: At5g0... 270 2e-71
gb|AAL12220.1| porphobilinogen deaminase [Triticum aestivum] 237 2e-61
dbj|BAB41183.1| porphobilinogen deaminase [Amaranthus tricolor] 181 1e-44
>sp|Q43082|HEM3_PEA Porphobilinogen deaminase, chloroplast precursor (PBG)
(Hydroxymethylbilane synthase) (HMBS)
(Pre-uroporphyrinogen synthase) gi|541971|pir||JQ2278
hydroxymethylbilane synthase (EC 4.3.1.8) precursor,
chloroplast - garden pea gi|313724|emb|CAA51820.1|
hydroxymethylbilane synthase [Pisum sativum]
Length = 369
Score = 289 bits (739), Expect = 4e-77
Identities = 142/163 (87%), Positives = 157/163 (96%)
Frame = -1
Query: 883 DNFRGNVQTRLKKLNEGVVKATLLALAGLKRLNMTENVTSTLSIEEMLPAVAQGAIGIAC 704
DNFRGNVQTRL+KL+EGVVKATLLALAGLKRLNMTENVTSTLSI++MLPAVAQGAIGIAC
Sbjct: 207 DNFRGNVQTRLRKLSEGVVKATLLALAGLKRLNMTENVTSTLSIDDMLPAVAQGAIGIAC 266
Query: 703 RSDDDKMAEYIASLNHEETRLAVVCERAFLLTLDGSCRTPIAGYASRNEDGNCLFRGLVA 524
RS+DDKMAEY+ASLNHEETRLA+ CERAFL TLDGSCRTPIAGYASR++DGNCLFRGLVA
Sbjct: 267 RSNDDKMAEYLASLNHEETRLAISCERAFLTTLDGSCRTPIAGYASRDKDGNCLFRGLVA 326
Query: 523 SPDGTRVLETSRVGPYAVEDMIEMGKDAGKELLSRAGPNFFSS 395
SPDGTRVLETSR+G Y EDM+++GKDAG+ELLSRAGP FF+S
Sbjct: 327 SPDGTRVLETSRIGSYTYEDMMKIGKDAGEELLSRAGPGFFNS 369
>gb|AAM64573.1| hydroxymethylbilane synthase [Arabidopsis thaliana]
Length = 382
Score = 270 bits (691), Expect = 2e-71
Identities = 132/163 (80%), Positives = 149/163 (90%)
Frame = -1
Query: 883 DNFRGNVQTRLKKLNEGVVKATLLALAGLKRLNMTENVTSTLSIEEMLPAVAQGAIGIAC 704
+NFRGNVQTRL KL G V+ATLLALAGLKRL+MTENV S LS++EMLPAVAQGAIGIAC
Sbjct: 220 ENFRGNVQTRLSKLQGGKVQATLLALAGLKRLSMTENVASILSLDEMLPAVAQGAIGIAC 279
Query: 703 RSDDDKMAEYIASLNHEETRLAVVCERAFLLTLDGSCRTPIAGYASRNEDGNCLFRGLVA 524
R+DDDKMA Y+ASLNHEETRLA+ CERAFL TLDGSCRTPIAGYAS++E+GNC+FRGLVA
Sbjct: 280 RTDDDKMATYLASLNHEETRLAISCERAFLETLDGSCRTPIAGYASKDEEGNCIFRGLVA 339
Query: 523 SPDGTRVLETSRVGPYAVEDMIEMGKDAGKELLSRAGPNFFSS 395
SPDGT+VLETSR GPY EDM++MGKDAG+ELLSRAGP FF +
Sbjct: 340 SPDGTKVLETSRKGPYVYEDMVKMGKDAGQELLSRAGPGFFGN 382
>ref|NP_196445.1| hydroxymethylbilane synthase; protein id: At5g08280.1, supported by
cDNA: 3051., supported by cDNA: gi_16930520, supported
by cDNA: gi_17979401 [Arabidopsis thaliana]
gi|2495179|sp|Q43316|HEM3_ARATH Porphobilinogen
deaminase, chloroplast precursor (PBG)
(Hydroxymethylbilane synthase) (HMBS)
(Pre-uroporphyrinogen synthase) gi|1084340|pir||S50762
hydroxymethylbilane synthase (EC 4.3.1.8) precursor -
Arabidopsis thaliana gi|313150|emb|CAA51941.1|
hydroxymethylbilane synthase [Arabidopsis thaliana]
gi|313838|emb|CAA52061.1| hydroxymethylbilane synthase
[Arabidopsis thaliana] gi|10178270|emb|CAC08328.1|
hydroxymethylbilane synthase [Arabidopsis thaliana]
gi|16930521|gb|AAL31946.1|AF419614_1 AT5g08280/F8L15_10
[Arabidopsis thaliana] gi|17979402|gb|AAL49926.1|
putative hydroxymethylbilane synthase [Arabidopsis
thaliana] gi|21689853|gb|AAM67570.1| putative
hydroxymethylbilane synthase [Arabidopsis thaliana]
Length = 382
Score = 270 bits (691), Expect = 2e-71
Identities = 132/163 (80%), Positives = 149/163 (90%)
Frame = -1
Query: 883 DNFRGNVQTRLKKLNEGVVKATLLALAGLKRLNMTENVTSTLSIEEMLPAVAQGAIGIAC 704
+NFRGNVQTRL KL G V+ATLLALAGLKRL+MTENV S LS++EMLPAVAQGAIGIAC
Sbjct: 220 ENFRGNVQTRLSKLQGGKVQATLLALAGLKRLSMTENVASILSLDEMLPAVAQGAIGIAC 279
Query: 703 RSDDDKMAEYIASLNHEETRLAVVCERAFLLTLDGSCRTPIAGYASRNEDGNCLFRGLVA 524
R+DDDKMA Y+ASLNHEETRLA+ CERAFL TLDGSCRTPIAGYAS++E+GNC+FRGLVA
Sbjct: 280 RTDDDKMATYLASLNHEETRLAISCERAFLETLDGSCRTPIAGYASKDEEGNCIFRGLVA 339
Query: 523 SPDGTRVLETSRVGPYAVEDMIEMGKDAGKELLSRAGPNFFSS 395
SPDGT+VLETSR GPY EDM++MGKDAG+ELLSRAGP FF +
Sbjct: 340 SPDGTKVLETSRKGPYVYEDMVKMGKDAGQELLSRAGPGFFGN 382
>gb|AAL12220.1| porphobilinogen deaminase [Triticum aestivum]
Length = 308
Score = 237 bits (605), Expect = 2e-61
Identities = 117/164 (71%), Positives = 135/164 (81%)
Frame = -1
Query: 880 NFRGNVQTRLKKLNEGVVKATLLALAGLKRLNMTENVTSTLSIEEMLPAVAQGAIGIACR 701
NFRGNVQTRL+KL EG V ATLLALAGLKRL M E TS LS++EMLPAVAQGAIGI CR
Sbjct: 145 NFRGNVQTRLRKLKEGDVHATLLALAGLKRLGMPETATSVLSVDEMLPAVAQGAIGITCR 204
Query: 700 SDDDKMAEYIASLNHEETRLAVVCERAFLLTLDGSCRTPIAGYASRNEDGNCLFRGLVAS 521
S+DDKM EY++SLNHE+TRLAV CER FL LDG+CRTPIA YA R++DGNC FRGL+AS
Sbjct: 205 SNDDKMMEYLSSLNHEDTRLAVACEREFLSVLDGNCRTPIAAYAYRDKDGNCSFRGLLAS 264
Query: 520 PDGTRVLETSRVGPYAVEDMIEMGKDAGKELLSRAGPNFFSS*Q 389
PDG+ V ETSR G Y+ +DM+ +G+DAG EL S+AGP FF Q
Sbjct: 265 PDGSIVYETSRSGTYSFDDMVALGQDAGHELKSKAGPGFFDGLQ 308
>dbj|BAB41183.1| porphobilinogen deaminase [Amaranthus tricolor]
Length = 198
Score = 181 bits (460), Expect = 1e-44
Identities = 90/105 (85%), Positives = 98/105 (92%)
Frame = -1
Query: 883 DNFRGNVQTRLKKLNEGVVKATLLALAGLKRLNMTENVTSTLSIEEMLPAVAQGAIGIAC 704
DNFRGNVQTRL+KLNEG+V+ATLLALAGLKRLNMTENV+S LSI++MLPAVAQGAIGIAC
Sbjct: 94 DNFRGNVQTRLRKLNEGLVQATLLALAGLKRLNMTENVSSVLSIDDMLPAVAQGAIGIAC 153
Query: 703 RSDDDKMAEYIASLNHEETRLAVVCERAFLLTLDGSCRTPIAGYA 569
DDDKMA Y+A LNHEET+LAV CERAFL TLDGSCRTPIAGYA
Sbjct: 154 LQDDDKMANYLALLNHEETQLAVACERAFLETLDGSCRTPIAGYA 198
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 763,708,424
Number of Sequences: 1393205
Number of extensions: 16630188
Number of successful extensions: 48119
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 45849
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 47975
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 47660818527
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)