Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
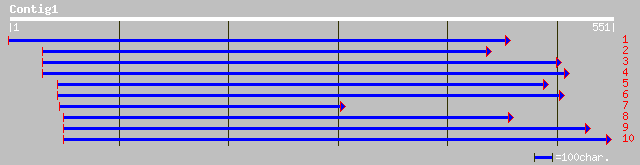
Query= KMC001948A_C01 KMC001948A_c01
(543 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_171839.1| putative 1-aminocyclopropane-1-carboxylate oxid... 146 1e-34
ref|NP_200761.1| 1-aminocyclopropane-1-carboxylate oxidase - lik... 144 5e-34
ref|NP_171930.1| putative 1-aminocyclopropane-1-carboxylate oxid... 142 3e-33
dbj|BAC19916.1| putative 1-aminocyclopropane-1-carboxylate oxida... 140 1e-32
ref|NP_200762.1| 1-aminocyclopropane-1-carboxylate oxidase - lik... 139 3e-32
>ref|NP_171839.1| putative 1-aminocyclopropane-1-carboxylate oxidase; protein id:
At1g03400.1, supported by cDNA: gi_15146295 [Arabidopsis
thaliana] gi|7433201|pir||T00917 hypothetical protein
F21B7.31 - Arabidopsis thaliana
gi|15146296|gb|AAK83631.1| At1g03400/F21B7_31
[Arabidopsis thaliana] gi|22137088|gb|AAM91389.1|
At1g03400/F21B7_31 [Arabidopsis thaliana]
Length = 351
Score = 146 bits (369), Expect = 1e-34
Identities = 72/137 (52%), Positives = 99/137 (71%), Gaps = 2/137 (1%)
Frame = -2
Query: 542 PQPDLTVGLNSHADPGALTVVLQDHIGGLQVRTTEGWVDVKPLDGALVINIGDLLQIISN 363
PQPDLT+G+N H D LTV+LQD++GGLQV + W+DV P+ GALVINIGD LQ+I+N
Sbjct: 213 PQPDLTIGINKHTDISFLTVLLQDNVGGLQVFHEQYWIDVTPVPGALVINIGDFLQLITN 272
Query: 362 EEYKSADHRVLANSANEPRVSVAVFLNPGNR--EKLFGPLPELTSAEKPALYRNFTLDEF 189
+++ SA+HRV+AN ++EPR SVA+ + R +++GP+ +L SAE PA YR+ TL EF
Sbjct: 273 DKFISAEHRVIANGSSEPRTSVAIVFSTFMRAYSRVYGPIKDLLSAENPAKYRDCTLTEF 332
Query: 188 LTRFFKKELDGKSLTNY 138
T F K LD L ++
Sbjct: 333 STIFSSKTLDAPKLHHF 349
>ref|NP_200761.1| 1-aminocyclopropane-1-carboxylate oxidase - like protein; protein
id: At5g59530.1 [Arabidopsis thaliana]
gi|8885557|dbj|BAA97487.1| leucoanthocyanidin
dioxygenase-like protein [Arabidopsis thaliana]
Length = 364
Score = 144 bits (364), Expect = 5e-34
Identities = 73/139 (52%), Positives = 99/139 (70%), Gaps = 2/139 (1%)
Frame = -2
Query: 542 PQPDLTVGLNSHADPGALTVVLQDHIGGLQVRTTEGWVDVKPLDGALVINIGDLLQIISN 363
PQPDLT+G++ H+D LTV+LQD+IGGLQ+ + WVDV PL GALV+N+GD LQ+I+N
Sbjct: 226 PQPDLTLGISKHSDNSFLTVLLQDNIGGLQILHQDSWVDVSPLPGALVVNVGDFLQLITN 285
Query: 362 EEYKSADHRVLANSANEPRVSVAVFLNPGNREK--LFGPLPELTSAEKPALYRNFTLDEF 189
+++ S +HRVLAN+ PR+SVA F + RE ++GP+ EL S E P YR+ TL E+
Sbjct: 286 DKFISVEHRVLANTRG-PRISVASFFSSSIRENSTVYGPMKELVSEENPPKYRDTTLREY 344
Query: 188 LTRFFKKELDGKSLTNYFR 132
+FKK LDG S + FR
Sbjct: 345 SEGYFKKGLDGTSHLSNFR 363
>ref|NP_171930.1| putative 1-aminocyclopropane-1-carboxylate oxidase; protein id:
At1g04350.1, supported by cDNA: gi_15292974, supported
by cDNA: gi_21281120 [Arabidopsis thaliana]
gi|25285693|pir||A86175 hypothetical protein [imported]
- Arabidopsis thaliana gi|1903360|gb|AAB70442.1| Similar
to Arabidopsis 2A6 (gb|X83096). EST gb|T76913 comes from
this gene. [Arabidopsis thaliana]
gi|15292975|gb|AAK93598.1| putative
1-aminocyclopropane-1-carboxylate oxidase [Arabidopsis
thaliana] gi|21281121|gb|AAM45017.1| putative
1-aminocyclopropane-1-carboxylate oxidase [Arabidopsis
thaliana] gi|23397116|gb|AAN31842.1| putative
1-aminocyclopropane-1-carboxylate oxidase [Arabidopsis
thaliana]
Length = 360
Score = 142 bits (358), Expect = 3e-33
Identities = 70/139 (50%), Positives = 96/139 (68%), Gaps = 2/139 (1%)
Frame = -2
Query: 542 PQPDLTVGLNSHADPGALTVVLQDHIGGLQVRTTEGWVDVKPLDGALVINIGDLLQIISN 363
PQPDLT+G N+H+D LT++LQD IGGLQ+ + WVDV P+ GALVIN+GD LQ+I+N
Sbjct: 221 PQPDLTIGTNNHSDNSFLTILLQDQIGGLQIFHQDCWVDVSPIPGALVINMGDFLQLITN 280
Query: 362 EEYKSADHRVLANSANEPRVSVAVFLNPGNR--EKLFGPLPELTSAEKPALYRNFTLDEF 189
++ S +HRVLAN A PR+SVA F + R ++GP+ EL S E P+ YR L E+
Sbjct: 281 DKVISVEHRVLANRAATPRISVASFFSTSMRPNSTVYGPIKELLSEENPSKYRVIDLKEY 340
Query: 188 LTRFFKKELDGKSLTNYFR 132
+FKK LDG S ++++
Sbjct: 341 TEGYFKKGLDGTSYLSHYK 359
>dbj|BAC19916.1| putative 1-aminocyclopropane-1-carboxylate oxidase [Oryza sativa
(japonica cultivar-group)]
Length = 396
Score = 140 bits (352), Expect = 1e-32
Identities = 73/147 (49%), Positives = 99/147 (66%), Gaps = 10/147 (6%)
Frame = -2
Query: 542 PQPDLTVGLNSHADPGALTVVLQDHIGGLQVRTTEG--------WVDVKPLDGALVINIG 387
P+P+ VG H DP TV+ QD +GGLQVR E WVDV P+ GALV+N+G
Sbjct: 247 PEPERVVGSLEHTDPSLFTVLAQDAVGGLQVRREEEEGGGGGGEWVDVAPVAGALVVNVG 306
Query: 386 DLLQIISNEEYKSADHRVLANSANEPRVSVAVFLNPGNRE--KLFGPLPELTSAEKPALY 213
D+L+++SNEEYKS +HRV+ S+ + RVS+AVF NP R+ LFGPLPEL +AE+PA +
Sbjct: 307 DVLKMVSNEEYKSVEHRVVIKSSQDARVSIAVFFNPAKRDASDLFGPLPELLTAERPARF 366
Query: 212 RNFTLDEFLTRFFKKELDGKSLTNYFR 132
R F++ EF+ R ++ GKS + FR
Sbjct: 367 RRFSVPEFM-RSRRESGHGKSSIDSFR 392
>ref|NP_200762.1| 1-aminocyclopropane-1-carboxylate oxidase - like protein; protein
id: At5g59540.1, supported by cDNA: gi_15983482
[Arabidopsis thaliana] gi|8885558|dbj|BAA97488.1|
leucoanthocyanidin dioxygenase-like protein [Arabidopsis
thaliana] gi|15983483|gb|AAL11609.1|AF424616_1
AT5g59540/f2o15_200 [Arabidopsis thaliana]
Length = 366
Score = 139 bits (349), Expect = 3e-32
Identities = 68/133 (51%), Positives = 94/133 (70%), Gaps = 2/133 (1%)
Frame = -2
Query: 542 PQPDLTVGLNSHADPGALTVVLQDHIGGLQVRTTEGWVDVKPLDGALVINIGDLLQIISN 363
PQPDLT+G+ H+D LT++LQD+IGGLQ+ + WVDV P+ GALV+NIGD LQ+I+N
Sbjct: 228 PQPDLTLGITKHSDNSFLTLLLQDNIGGLQILHQDSWVDVSPIHGALVVNIGDFLQLITN 287
Query: 362 EEYKSADHRVLANSANEPRVSVAVFLNPGNR--EKLFGPLPELTSAEKPALYRNFTLDEF 189
+++ S +HRVLAN PR+SVA F + R +++GP+ EL S E P YR+ T+ E+
Sbjct: 288 DKFVSVEHRVLANRQG-PRISVASFFSSSMRPNSRVYGPMKELVSEENPPKYRDITIKEY 346
Query: 188 LTRFFKKELDGKS 150
FF+K LDG S
Sbjct: 347 SKIFFEKGLDGTS 359
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 465,775,796
Number of Sequences: 1393205
Number of extensions: 9737453
Number of successful extensions: 25905
Number of sequences better than 10.0: 658
Number of HSP's better than 10.0 without gapping: 24023
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25294
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18750593680
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)