Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001938A_C01 KMC001938A_c01
(804 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
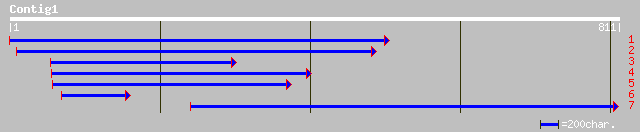
ref|NP_563837.1| Expressed protein; protein id: At1g09155.1, sup... 91 1e-25
pir||G86223 hypothetical protein [imported] - Arabidopsis thalia... 91 1e-25
gb|AAM64609.1| phloem-specific lectin PP2-like protein [Arabidop... 91 1e-25
ref|NP_176020.1| hypothetical protein; protein id: At1g56240.1 [... 86 1e-21
ref|NP_176021.1| hypothetical protein; protein id: At1g56250.1 [... 82 3e-21
>ref|NP_563837.1| Expressed protein; protein id: At1g09155.1, supported by cDNA:
31022. [Arabidopsis thaliana]
Length = 289
Score = 90.9 bits (224), Expect(2) = 1e-25
Identities = 59/129 (45%), Positives = 76/129 (58%), Gaps = 5/129 (3%)
Frame = -1
Query: 570 LIMNVSHRGYGLDSAPSEVSSTVANNVMHTKKVYL-WNKNEKNMCEKYTLFRGFHRDVAR 394
LIM V+ R YGLD P+E S V N K YL N+K E+ +F G R+
Sbjct: 157 LIMKVTSRAYGLDLVPAETSIKVGNGEKKIKSTYLSCLDNKKQQMER--VFYG-QREQRM 213
Query: 393 ETLIIQDQINPGPLKRDDGWMEIEVGEFFC---DGEIDEEVKVSVMEV-GYQLKGGLIVE 226
T + P RDDGWMEIE+GEF +G+ D+EV +S+ EV GYQLKGG+ ++
Sbjct: 214 ATHEVVRSHRREPEVRDDGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAID 273
Query: 225 GIEVRPKPV 199
GIEVRPKP+
Sbjct: 274 GIEVRPKPL 282
Score = 48.1 bits (113), Expect(2) = 1e-25
Identities = 22/35 (62%), Positives = 28/35 (79%)
Frame = -2
Query: 668 QIFKLDKSSGKKSYILSARELSITWSSDPLYWTSS 564
+IFK++K SGK SYILS+R+LSITWS YW+ S
Sbjct: 83 KIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWS 117
>pir||G86223 hypothetical protein [imported] - Arabidopsis thaliana
gi|3249107|gb|AAC24090.1| Contains similarity to
phloem-specific lectin PP2 gb|Z17331 from Cucubita
maxima. [Arabidopsis thaliana]
Length = 288
Score = 90.9 bits (224), Expect(2) = 1e-25
Identities = 59/129 (45%), Positives = 76/129 (58%), Gaps = 5/129 (3%)
Frame = -1
Query: 570 LIMNVSHRGYGLDSAPSEVSSTVANNVMHTKKVYL-WNKNEKNMCEKYTLFRGFHRDVAR 394
LIM V+ R YGLD P+E S V N K YL N+K E+ +F G R+
Sbjct: 156 LIMKVTSRAYGLDLVPAETSIKVGNGEKKIKSTYLSCLDNKKQQMER--VFYG-QREQRM 212
Query: 393 ETLIIQDQINPGPLKRDDGWMEIEVGEFFC---DGEIDEEVKVSVMEV-GYQLKGGLIVE 226
T + P RDDGWMEIE+GEF +G+ D+EV +S+ EV GYQLKGG+ ++
Sbjct: 213 ATHEVVRSHRREPEVRDDGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAID 272
Query: 225 GIEVRPKPV 199
GIEVRPKP+
Sbjct: 273 GIEVRPKPL 281
Score = 48.1 bits (113), Expect(2) = 1e-25
Identities = 22/35 (62%), Positives = 28/35 (79%)
Frame = -2
Query: 668 QIFKLDKSSGKKSYILSARELSITWSSDPLYWTSS 564
+IFK++K SGK SYILS+R+LSITWS YW+ S
Sbjct: 82 KIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWS 116
>gb|AAM64609.1| phloem-specific lectin PP2-like protein [Arabidopsis thaliana]
Length = 288
Score = 90.9 bits (224), Expect(2) = 1e-25
Identities = 59/129 (45%), Positives = 76/129 (58%), Gaps = 5/129 (3%)
Frame = -1
Query: 570 LIMNVSHRGYGLDSAPSEVSSTVANNVMHTKKVYL-WNKNEKNMCEKYTLFRGFHRDVAR 394
LIM V+ R YGLD P+E S V N K YL N+K E+ +F G R+
Sbjct: 156 LIMKVTSRAYGLDLVPAETSIKVGNGEKKIKSTYLSCLDNKKQQMER--VFYG-QREQRM 212
Query: 393 ETLIIQDQINPGPLKRDDGWMEIEVGEFFC---DGEIDEEVKVSVMEV-GYQLKGGLIVE 226
T + P RDDGWMEIE+GEF +G+ D+EV +S+ EV GYQLKGG+ ++
Sbjct: 213 ATHEVVRSHRREPEVRDDGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAID 272
Query: 225 GIEVRPKPV 199
GIEVRPKP+
Sbjct: 273 GIEVRPKPL 281
Score = 48.1 bits (113), Expect(2) = 1e-25
Identities = 22/35 (62%), Positives = 28/35 (79%)
Frame = -2
Query: 668 QIFKLDKSSGKKSYILSARELSITWSSDPLYWTSS 564
+IFK++K SGK SYILS+R+LSITWS YW+ S
Sbjct: 82 KIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWS 116
>ref|NP_176020.1| hypothetical protein; protein id: At1g56240.1 [Arabidopsis
thaliana] gi|25366251|pir||A96604 hypothetical protein
F14G9.15 [imported] - Arabidopsis thaliana
gi|12321750|gb|AAG50910.1|AC069159_11 hypothetical
protein [Arabidopsis thaliana]
Length = 284
Score = 85.5 bits (210), Expect(2) = 1e-21
Identities = 54/127 (42%), Positives = 72/127 (56%), Gaps = 3/127 (2%)
Frame = -1
Query: 570 LIMNVSHRGYGLDSAPSEVSSTVANNVMHTKKVYLWNKNEKNMCEKYTLFRGFHRDVARE 391
LIM V++ YGLD P+E S N + YL +EK K LF G +
Sbjct: 158 LIMKVTNGAYGLDLVPAETSVKSKNGQNNKNTTYLCCLDEKKQQMK-RLFYGNREERMAM 216
Query: 390 TL--IIQDQINPGPLKRDDGWMEIEVGEFFCDGEIDEEVKVSVMEV-GYQLKGGLIVEGI 220
T+ + D P RDDGW+EIE+GEF D+EV +S+ EV GYQLKGG++++GI
Sbjct: 217 TVEAVGGDGKRREPKARDDGWLEIELGEFVTREGEDDEVNMSLTEVKGYQLKGGIVIDGI 276
Query: 219 EVRPKPV 199
EVRP P+
Sbjct: 277 EVRPIPL 283
Score = 40.4 bits (93), Expect(2) = 1e-21
Identities = 29/78 (37%), Positives = 44/78 (56%), Gaps = 6/78 (7%)
Frame = -2
Query: 770 FFPSFISCLISKN----RTFSLINLLFMLYI--YVVP*HMQIFKLDKSSGKKSYILSARE 609
F PS LIS++ R FS ++ ++ ++FK++K SGK SYILSAR+
Sbjct: 44 FLPSHYKSLISQSTDHHRIFSSKKEIYRCLCDSLLIDNARKLFKINKFSGKISYILSARD 103
Query: 608 LSITWSSDPLYWTSS*MS 555
+SIT+S Y + S +S
Sbjct: 104 ISITYSDHASYCSWSNVS 121
>ref|NP_176021.1| hypothetical protein; protein id: At1g56250.1 [Arabidopsis
thaliana] gi|25366254|pir||B96604 hypothetical protein
F14G9.14 [imported] - Arabidopsis thaliana
gi|12321747|gb|AAG50907.1|AC069159_8 hypothetical
protein [Arabidopsis thaliana]
Length = 282
Score = 81.6 bits (200), Expect(2) = 3e-21
Identities = 53/125 (42%), Positives = 70/125 (55%), Gaps = 3/125 (2%)
Frame = -1
Query: 570 LIMNVSHRGYGLDSAPSEVSSTVANNVMHTKKVYLWNKNEKNMCEKYTLFRGFHRDVARE 391
LI+ V+ YGLD P+E S N + YL +EK K LF G +
Sbjct: 158 LIVKVTKGAYGLDLVPAETSIKSKNGQISKSATYLCCLDEKKQQMK-RLFYGNREERMAM 216
Query: 390 TL--IIQDQINPGPLKRDDGWMEIEVGEFFCDGEIDEEVKVSVMEV-GYQLKGGLIVEGI 220
T+ + D P RDDGWMEIE+GEF D+EV +++ EV GYQLKGG++++GI
Sbjct: 217 TVEAVGGDGKRREPKCRDDGWMEIELGEFETREGEDDEVNMTLTEVKGYQLKGGILIDGI 276
Query: 219 EVRPK 205
EVRPK
Sbjct: 277 EVRPK 281
Score = 42.7 bits (99), Expect(2) = 3e-21
Identities = 20/38 (52%), Positives = 29/38 (75%)
Frame = -2
Query: 668 QIFKLDKSSGKKSYILSARELSITWSSDPLYWTSS*MS 555
++FK++K SGK SY+LSAR++SIT S YW+ S +S
Sbjct: 84 KLFKINKFSGKISYVLSARDISITHSDHASYWSWSNVS 121
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 694,589,658
Number of Sequences: 1393205
Number of extensions: 15583515
Number of successful extensions: 42902
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 39068
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42473
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40896270532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)