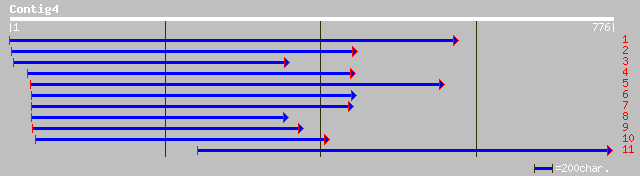
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001925A_C04 KMC001925A_c04
(772 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_566986.1| chloroplast lumen common protein family; protei... 162 4e-39
pir||T45896 hypothetical protein F4P12.260 - Arabidopsis thalian... 162 4e-39
ref|NP_565860.1| chloroplast lumen common protein family; protei... 161 8e-39
gb|AAM66986.1| unknown [Arabidopsis thaliana] 160 2e-38
pir||T48280 hypothetical protein T22P11.180 - Arabidopsis thalia... 154 2e-36
>ref|NP_566986.1| chloroplast lumen common protein family; protein id: At3g53560.1,
supported by cDNA: 36212., supported by cDNA:
gi_20466551 [Arabidopsis thaliana]
gi|20466552|gb|AAM20593.1| putative protein [Arabidopsis
thaliana] gi|23198132|gb|AAN15593.1| putative protein
[Arabidopsis thaliana]
Length = 340
Score = 162 bits (411), Expect = 4e-39
Identities = 80/135 (59%), Positives = 107/135 (79%), Gaps = 3/135 (2%)
Frame = -1
Query: 772 RDPFDAEAYHGLLKATSELNEPIGDLLKRIEEAMKHCEAEKDKAFEAREFKLLIAQIKVI 593
+DP EAYHGLL A S+ + ++ RIEEAM C+ E ++ + R+FKLL+AQI+VI
Sbjct: 193 KDPLRVEAYHGLLMAYSDAGLDLKEVESRIEEAMLKCKKENNQN-DFRDFKLLVAQIRVI 251
Query: 592 EDDYSGALKVYQELVKEEPKDFRPYLCQGVVYTLLKKKDEADKQFAMYRKLLPKDHPYQE 413
E +S ALK+YQELVKEEP+DFRPYLCQG++YTLLKKKD+A++QF +RKL+PK+HPY+E
Sbjct: 252 EGKHSEALKLYQELVKEEPRDFRPYLCQGIIYTLLKKKDKAEEQFDNFRKLVPKNHPYRE 311
Query: 412 YFEDN---TQVFSQK 377
YF DN T++FS+K
Sbjct: 312 YFMDNMIATKLFSEK 326
>pir||T45896 hypothetical protein F4P12.260 - Arabidopsis thaliana
gi|6729507|emb|CAB67663.1| putative protein [Arabidopsis
thaliana]
Length = 388
Score = 162 bits (411), Expect = 4e-39
Identities = 80/135 (59%), Positives = 107/135 (79%), Gaps = 3/135 (2%)
Frame = -1
Query: 772 RDPFDAEAYHGLLKATSELNEPIGDLLKRIEEAMKHCEAEKDKAFEAREFKLLIAQIKVI 593
+DP EAYHGLL A S+ + ++ RIEEAM C+ E ++ + R+FKLL+AQI+VI
Sbjct: 241 KDPLRVEAYHGLLMAYSDAGLDLKEVESRIEEAMLKCKKENNQN-DFRDFKLLVAQIRVI 299
Query: 592 EDDYSGALKVYQELVKEEPKDFRPYLCQGVVYTLLKKKDEADKQFAMYRKLLPKDHPYQE 413
E +S ALK+YQELVKEEP+DFRPYLCQG++YTLLKKKD+A++QF +RKL+PK+HPY+E
Sbjct: 300 EGKHSEALKLYQELVKEEPRDFRPYLCQGIIYTLLKKKDKAEEQFDNFRKLVPKNHPYRE 359
Query: 412 YFEDN---TQVFSQK 377
YF DN T++FS+K
Sbjct: 360 YFMDNMIATKLFSEK 374
>ref|NP_565860.1| chloroplast lumen common protein family; protein id: At2g37400.1,
supported by cDNA: 9001. [Arabidopsis thaliana]
gi|25408548|pir||C84792 hypothetical protein At2g37400
[imported] - Arabidopsis thaliana
gi|4056493|gb|AAC98059.1| chloroplast lumen common
protein family [Arabidopsis thaliana]
Length = 333
Score = 161 bits (408), Expect = 8e-39
Identities = 75/137 (54%), Positives = 111/137 (80%), Gaps = 3/137 (2%)
Frame = -1
Query: 772 RDPFDAEAYHGLLKATSELNEPIGDLLKRIEEAMKHCEAEKDKAFEAREFKLLIAQIKVI 593
+DP EAYHGL+ A S+ + + + KRIEEAM C+ EK++ + R+FKLL+AQI+VI
Sbjct: 193 KDPLRVEAYHGLVMAYSDSGDDLNAVEKRIEEAMVRCKKEKNRK-DLRDFKLLVAQIRVI 251
Query: 592 EDDYSGALKVYQELVKEEPKDFRPYLCQGVVYTLLKKKDEADKQFAMYRKLLPKDHPYQE 413
E ++ ALK+Y+ELVKEEP+DFRPYLCQG++YT+LKK++EA+KQF +R+L+PK+HPY+E
Sbjct: 252 EGKHNEALKLYEELVKEEPRDFRPYLCQGIIYTVLKKENEAEKQFEKFRRLVPKNHPYRE 311
Query: 412 YFEDN---TQVFSQKLR 371
YF DN +++F++K++
Sbjct: 312 YFMDNMVASKLFAEKVQ 328
>gb|AAM66986.1| unknown [Arabidopsis thaliana]
Length = 333
Score = 160 bits (404), Expect = 2e-38
Identities = 74/137 (54%), Positives = 111/137 (81%), Gaps = 3/137 (2%)
Frame = -1
Query: 772 RDPFDAEAYHGLLKATSELNEPIGDLLKRIEEAMKHCEAEKDKAFEAREFKLLIAQIKVI 593
+DP EAYHGL+ A S+ + + + +RIEEAM C+ EK++ + R+FKLL+AQI+VI
Sbjct: 193 KDPLRVEAYHGLVMAYSDSGDDLNAVEQRIEEAMVRCKKEKNRK-DLRDFKLLVAQIRVI 251
Query: 592 EDDYSGALKVYQELVKEEPKDFRPYLCQGVVYTLLKKKDEADKQFAMYRKLLPKDHPYQE 413
E ++ ALK+Y+ELVKEEP+DFRPYLCQG++YT+LKK++EA+KQF +R+L+PK+HPY+E
Sbjct: 252 EGKHNEALKLYEELVKEEPRDFRPYLCQGIIYTVLKKENEAEKQFEKFRRLVPKNHPYRE 311
Query: 412 YFEDN---TQVFSQKLR 371
YF DN +++F++K++
Sbjct: 312 YFMDNMVASKLFAEKVQ 328
>pir||T48280 hypothetical protein T22P11.180 - Arabidopsis thaliana
gi|7413648|emb|CAB85996.1| putative protein [Arabidopsis
thaliana]
Length = 407
Score = 154 bits (388), Expect = 2e-36
Identities = 73/125 (58%), Positives = 98/125 (78%)
Frame = -1
Query: 772 RDPFDAEAYHGLLKATSELNEPIGDLLKRIEEAMKHCEAEKDKAFEAREFKLLIAQIKVI 593
+DPF EAYHGL+ A SE + ++ RI EA++ C+ E K F R+F LLIAQI+VI
Sbjct: 192 KDPFRVEAYHGLVMAYSESESKLSEIESRINEAIEKCKKENKKDF--RDFMLLIAQIRVI 249
Query: 592 EDDYSGALKVYQELVKEEPKDFRPYLCQGVVYTLLKKKDEADKQFAMYRKLLPKDHPYQE 413
+ + AL+VYQELVK+EPKDFRPYLCQG++YTL+KKKDEA+KQFA +R+L+P++HPY+E
Sbjct: 250 KGNPIEALRVYQELVKDEPKDFRPYLCQGLIYTLMKKKDEAEKQFAEFRRLVPENHPYKE 309
Query: 412 YFEDN 398
Y + N
Sbjct: 310 YLDAN 314
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 572,633,728
Number of Sequences: 1393205
Number of extensions: 11729192
Number of successful extensions: 45929
Number of sequences better than 10.0: 333
Number of HSP's better than 10.0 without gapping: 39950
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44725
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37815044670
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)