Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001924A_C02 KMC001924A_c02
(630 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
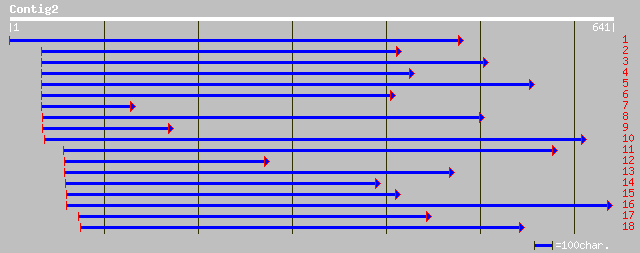
ref|NP_175413.1| mitotic checkpoint protein, putative; protein i... 278 3e-74
ref|NP_566644.1| mitotic checkpoint protein, putative; protein i... 278 6e-74
dbj|BAC57743.1| putative mitotic checkpoint protein [Oryza sativ... 216 2e-55
gb|AAO66553.1| putative mitotic checkpoint protein, 5'-partial [... 200 1e-50
ref|NP_004716.1| BUB3 budding uninhibited by benzimidazoles 3 ho... 170 2e-41
>ref|NP_175413.1| mitotic checkpoint protein, putative; protein id: At1g49910.1
[Arabidopsis thaliana]
gi|12597855|gb|AAG60165.1|AC074110_3 mitotic checkpoint
protein, putative [Arabidopsis thaliana]
Length = 339
Score = 278 bits (712), Expect = 3e-74
Identities = 131/144 (90%), Positives = 139/144 (95%)
Frame = -2
Query: 629 YPNGTGYALSSVEGRVAMEFFDLSEASQAKKYAFKCHRKSEAGRDFVYPVNAMAFHPIYG 450
YPNGTGYALSSVEGRV+MEFFDLSEA+QAKKYAFKCHRKSE GRD VYPVNA+AFHPIYG
Sbjct: 194 YPNGTGYALSSVEGRVSMEFFDLSEAAQAKKYAFKCHRKSEDGRDIVYPVNAIAFHPIYG 253
Query: 449 TFATGGCDGYVNVWDGNNKKRLYQYSKYPTSIAALSFSRDGRLLAVASSYTFEEGPKPHE 270
TFA+GGCDG+VN+WDGNNKKRLYQYSKYPTSIAALSFSRDG LLAVASSYTFEEG KPHE
Sbjct: 254 TFASGGCDGFVNIWDGNNKKRLYQYSKYPTSIAALSFSRDGGLLAVASSYTFEEGDKPHE 313
Query: 269 KDAIYVRSVNEIEVKPKPQVYPNP 198
DAI+VRSVNEIEVKPKP+VYPNP
Sbjct: 314 PDAIFVRSVNEIEVKPKPKVYPNP 337
>ref|NP_566644.1| mitotic checkpoint protein, putative; protein id: At3g19590.1,
supported by cDNA: 35031. [Arabidopsis thaliana]
gi|25455792|pir||T52386 mitotic checkpoint protein
[imported] - Arabidopsis thaliana
gi|9294423|dbj|BAB02543.1| mitotic checkpoint protein
[Arabidopsis thaliana] gi|21593004|gb|AAM64953.1|
mitotic checkpoint protein, putative [Arabidopsis
thaliana] gi|28393726|gb|AAO42274.1| putative mitotic
checkpoint protein [Arabidopsis thaliana]
Length = 340
Score = 278 bits (710), Expect = 6e-74
Identities = 131/146 (89%), Positives = 141/146 (95%)
Frame = -2
Query: 629 YPNGTGYALSSVEGRVAMEFFDLSEASQAKKYAFKCHRKSEAGRDFVYPVNAMAFHPIYG 450
YPNGTGYALSSVEGRVAMEFFDLSEA+QAKKYAFKCHRKSEAGRD VYPVN++AFHPIYG
Sbjct: 195 YPNGTGYALSSVEGRVAMEFFDLSEAAQAKKYAFKCHRKSEAGRDIVYPVNSIAFHPIYG 254
Query: 449 TFATGGCDGYVNVWDGNNKKRLYQYSKYPTSIAALSFSRDGRLLAVASSYTFEEGPKPHE 270
TFATGGCDG+VN+WDGNNKKRLYQYSKYPTSI+ALSFSRDG+LLAVASSYTFEEG K E
Sbjct: 255 TFATGGCDGFVNIWDGNNKKRLYQYSKYPTSISALSFSRDGQLLAVASSYTFEEGEKSQE 314
Query: 269 KDAIYVRSVNEIEVKPKPQVYPNPAA 192
+AI+VRSVNEIEVKPKP+VYPNPAA
Sbjct: 315 PEAIFVRSVNEIEVKPKPKVYPNPAA 340
>dbj|BAC57743.1| putative mitotic checkpoint protein [Oryza sativa (japonica
cultivar-group)]
Length = 364
Score = 216 bits (551), Expect = 2e-55
Identities = 96/140 (68%), Positives = 122/140 (86%)
Frame = -2
Query: 629 YPNGTGYALSSVEGRVAMEFFDLSEASQAKKYAFKCHRKSEAGRDFVYPVNAMAFHPIYG 450
YPNGTG+AL SVEGRVAMEF+D SE++ KKY+FKCHR E G VYPVNA++FHP++G
Sbjct: 212 YPNGTGFALGSVEGRVAMEFYDQSESAPYKKYSFKCHRVPEDGETKVYPVNAISFHPVHG 271
Query: 449 TFATGGCDGYVNVWDGNNKKRLYQYSKYPTSIAALSFSRDGRLLAVASSYTFEEGPKPHE 270
TFATGGCD +VN+WDG N+++L+Q+ +YP+SIAALSFSRDGRLLAVASSYT+EEG PH
Sbjct: 272 TFATGGCDRFVNLWDGANRRKLFQFPRYPSSIAALSFSRDGRLLAVASSYTYEEGDIPHP 331
Query: 269 KDAIYVRSVNEIEVKPKPQV 210
DAI++R VNE++VKP+P++
Sbjct: 332 PDAIFIRDVNEVQVKPRPKI 351
>gb|AAO66553.1| putative mitotic checkpoint protein, 5'-partial [Oryza sativa
(japonica cultivar-group)]
Length = 113
Score = 200 bits (508), Expect = 1e-50
Identities = 93/113 (82%), Positives = 102/113 (89%)
Frame = -2
Query: 536 YAFKCHRKSEAGRDFVYPVNAMAFHPIYGTFATGGCDGYVNVWDGNNKKRLYQYSKYPTS 357
YAFKCHRKSEAGRD VYPVNA+AFHPIYGTFATGGCDG+VNVWDG NKKRLYQYSKY +S
Sbjct: 1 YAFKCHRKSEAGRDTVYPVNAIAFHPIYGTFATGGCDGFVNVWDGINKKRLYQYSKYASS 60
Query: 356 IAALSFSRDGRLLAVASSYTFEEGPKPHEKDAIYVRSVNEIEVKPKPQVYPNP 198
IAALSFS+DG LLAVASSYT+EEG K HE DAI++RSVNE+EVKPKP+ P
Sbjct: 61 IAALSFSKDGHLLAVASSYTYEEGEKSHEPDAIFIRSVNEVEVKPKPKALAAP 113
>ref|NP_004716.1| BUB3 budding uninhibited by benzimidazoles 3 homolog; mitotic
checkpoint component; BUB3 (budding uninhibited by
benzimidazoles 3, yeast) homolog; budding uninhibited by
benomyl [Homo sapiens] gi|7387554|sp|O43684|BUB3_HUMAN
Mitotic checkpoint protein BUB3
gi|2921873|gb|AAC28438.1| spleen mitotic checkpoint BUB3
[Homo sapiens] gi|2981231|gb|AAC06258.1| mitotic
checkpoint component Bub3 [Homo sapiens]
gi|3639060|gb|AAC36307.1| kinetochore protein BUB3 [Homo
sapiens] gi|13477327|gb|AAH05138.1|AAH05138 BUB3
(budding uninhibited by benzimidazoles 3, yeast) homolog
[Homo sapiens] gi|18490881|gb|AAH22438.1| BUB3 budding
uninhibited by benzimidazoles 3 homolog (yeast) [Homo
sapiens]
Length = 328
Score = 170 bits (430), Expect = 2e-41
Identities = 79/137 (57%), Positives = 100/137 (72%)
Frame = -2
Query: 629 YPNGTGYALSSVEGRVAMEFFDLSEASQAKKYAFKCHRKSEAGRDFVYPVNAMAFHPIYG 450
+PN GY LSS+EGRVA+E+ D S Q KKYAFKCHR E + +YPVNA++FH I+
Sbjct: 188 FPNKQGYVLSSIEGRVAVEYLDPSPEVQKKKYAFKCHRLKENNIEQIYPVNAISFHNIHN 247
Query: 449 TFATGGCDGYVNVWDGNNKKRLYQYSKYPTSIAALSFSRDGRLLAVASSYTFEEGPKPHE 270
TFATGG DG+VN+WD NKKRL Q+ +YPTSIA+L+FS DG LA+ASSY +E H
Sbjct: 248 TFATGGSDGFVNIWDPFNKKRLCQFHRYPTSIASLAFSNDGTTLAIASSYMYEMDDTEHP 307
Query: 269 KDAIYVRSVNEIEVKPK 219
+D I++R V + E KPK
Sbjct: 308 EDGIFIRQVTDAETKPK 324
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 529,189,165
Number of Sequences: 1393205
Number of extensions: 11427375
Number of successful extensions: 30433
Number of sequences better than 10.0: 568
Number of HSP's better than 10.0 without gapping: 27422
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30256
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)