Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001920A_C01 KMC001920A_c01
(597 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
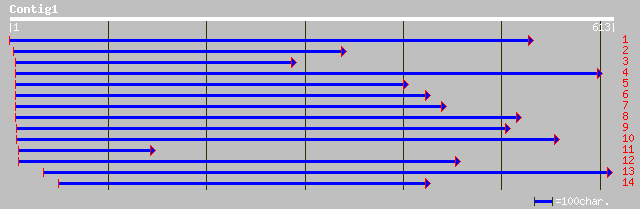
gb|AAG31636.1| endo-beta-1,4-D-glucanase [Lycopersicon esculentum] 54 1e-06
pir||T09873 probable cellulase (EC 3.2.1.4) - upland cotton (fra... 53 3e-06
dbj|BAC22691.1| endo-1,4-beta-D-glucanase [Pyrus communis] 49 6e-05
emb|CAC94006.1| endo-beta-1,4-glucanase [Fragaria x ananassa] 48 8e-05
gb|AAC78298.2| cellulase [Fragaria x ananassa] 48 8e-05
>gb|AAG31636.1| endo-beta-1,4-D-glucanase [Lycopersicon esculentum]
Length = 43
Score = 54.3 bits (129), Expect = 1e-06
Identities = 25/34 (73%), Positives = 28/34 (81%)
Frame = -2
Query: 596 PSWISSLSAGKSLEFVYIHAANSPADVSVAKYLL 495
P+WI+SL GKSLEFVYIH ANSPA VSV+ Y L
Sbjct: 9 PAWINSLPTGKSLEFVYIHTANSPAVVSVSSYTL 42
>pir||T09873 probable cellulase (EC 3.2.1.4) - upland cotton (fragment)
gi|2244740|dbj|BAA21111.1| endo-1,4-beta-glucanase
[Gossypium hirsutum]
Length = 324
Score = 52.8 bits (125), Expect = 3e-06
Identities = 26/35 (74%), Positives = 30/35 (85%)
Frame = -2
Query: 596 PSWISSLSAGKSLEFVYIHAANSPADVSVAKYLLA 492
P W++SL AGKSLEFVYIHAA SPADVSV+ Y +A
Sbjct: 291 PEWLNSLPAGKSLEFVYIHAA-SPADVSVSSYNIA 324
>dbj|BAC22691.1| endo-1,4-beta-D-glucanase [Pyrus communis]
Length = 620
Score = 48.5 bits (114), Expect = 6e-05
Identities = 24/35 (68%), Positives = 29/35 (82%)
Frame = -2
Query: 596 PSWISSLSAGKSLEFVYIHAANSPADVSVAKYLLA 492
PSWI+SL GKS+EFVYIH+A SPA+V V+ Y LA
Sbjct: 587 PSWINSLPVGKSMEFVYIHSA-SPANVLVSSYSLA 620
>emb|CAC94006.1| endo-beta-1,4-glucanase [Fragaria x ananassa]
Length = 620
Score = 48.1 bits (113), Expect = 8e-05
Identities = 25/35 (71%), Positives = 29/35 (82%)
Frame = -2
Query: 596 PSWISSLSAGKSLEFVYIHAANSPADVSVAKYLLA 492
PSW++SL AGKSLEFVYIHAA S A+V V+ Y LA
Sbjct: 587 PSWLNSLPAGKSLEFVYIHAA-SAANVLVSSYSLA 620
>gb|AAC78298.2| cellulase [Fragaria x ananassa]
Length = 620
Score = 48.1 bits (113), Expect = 8e-05
Identities = 25/35 (71%), Positives = 29/35 (82%)
Frame = -2
Query: 596 PSWISSLSAGKSLEFVYIHAANSPADVSVAKYLLA 492
PSW++SL AGKSLEFVYIHAA S A+V V+ Y LA
Sbjct: 587 PSWLNSLPAGKSLEFVYIHAA-SAANVLVSSYSLA 620
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 499,109,396
Number of Sequences: 1393205
Number of extensions: 10511086
Number of successful extensions: 24683
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 23985
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24677
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)