Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001918A_C01 KMC001918A_c01
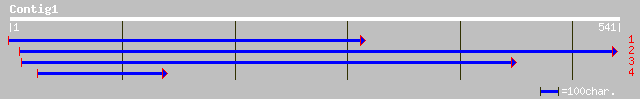
(540 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P93164|GGH_SOYBN GAMMA-GLUTAMYL HYDROLASE PRECURSOR (GAMMA-GL... 205 2e-52
sp|O65355|GGH_ARATH Gamma-glutamyl hydrolase precursor (Gamma-Gl... 202 3e-51
ref|NP_565186.1| gamma glutamyl hydrolase, putative; protein id:... 201 6e-51
ref|NP_177987.1| gamma glutamyl hydrolase, putative; protein id:... 198 4e-50
gb|AAM65646.1| gamma glutamyl hydrolase, putative [Arabidopsis t... 176 2e-43
>sp|P93164|GGH_SOYBN GAMMA-GLUTAMYL HYDROLASE PRECURSOR (GAMMA-GLU-X CARBOXYPEPTIDASE)
(CONJUGASE) (GH) gi|7488702|pir||T08837 probable
gamma-glutamyl hydrolase - soybean
gi|1679658|gb|AAB26960.1| gamma glutamyl hydrolase
[Glycine max]
Length = 342
Score = 205 bits (522), Expect = 2e-52
Identities = 105/165 (63%), Positives = 124/165 (74%), Gaps = 1/165 (0%)
Frame = +1
Query: 49 MSKDAVWVLFL-VTSFQCLLLSAETHPAILLPNHYNRNTPVTCPAPNPNLYYQPVIGIVT 225
M D+V LF VT F CLL + I LP+ + + V+C A +P+L Y+PVIGI+T
Sbjct: 1 MPNDSVLSLFFFVTLFTCLLSATSHDDHIFLPSQLHDDDSVSCTATDPSLNYKPVIGILT 60
Query: 226 HPGDGATGRLSNATGASYIAASYVKFVEAAGARVVPLIYNEPTEKLFKKLELVNGVLFTG 405
HPGDGA+GRLSNATG SYIAASYVKFVE+ GARV+PLIYNE E L KKL+LVNGVLFTG
Sbjct: 61 HPGDGASGRLSNATGVSYIAASYVKFVESGGARVIPLIYNESPENLNKKLDLVNGVLFTG 120
Query: 406 GWAKSGLYFETIGRIFKKALEKNDAGDHFPIYAICLGFELITMIV 540
GWA SG Y +T+G IFKKALE+NDAGDHFP+ A LG L+ IV
Sbjct: 121 GWAVSGPYLDTLGNIFKKALERNDAGDHFPVIAFNLGGNLVIRIV 165
>sp|O65355|GGH_ARATH Gamma-glutamyl hydrolase precursor (Gamma-Glu-X carboxypeptidase)
(Conjugase) (GH) gi|17979073|gb|AAL49804.1| putative
gamma glutamyl hydrolase [Arabidopsis thaliana]
gi|20465329|gb|AAM20068.1| putative gamma glutamyl
hydrolase [Arabidopsis thaliana]
Length = 347
Score = 202 bits (513), Expect = 3e-51
Identities = 101/162 (62%), Positives = 128/162 (78%), Gaps = 3/162 (1%)
Frame = +1
Query: 64 VWV-LFLVTSFQCLLLSAETHPAILLPNH--YNRNTPVTCPAPNPNLYYQPVIGIVTHPG 234
VW+ L ++ F+ ++ A+ ILLP+ ++ + C AP+PNL Y+PVIGI++HPG
Sbjct: 5 VWLPLVALSLFKDSIIMAKA-ATILLPSQTGFDISRSPVCSAPDPNLNYRPVIGILSHPG 63
Query: 235 DGATGRLSNATGASYIAASYVKFVEAAGARVVPLIYNEPTEKLFKKLELVNGVLFTGGWA 414
DGA+GRLSNAT AS IAASYVK E+ GARV+PLI+NEP E LF+KLELVNGV+ TGGWA
Sbjct: 64 DGASGRLSNATDASSIAASYVKLAESGGARVIPLIFNEPEEILFQKLELVNGVILTGGWA 123
Query: 415 KSGLYFETIGRIFKKALEKNDAGDHFPIYAICLGFELITMIV 540
K GLYFE + +IF K LE+NDAG+HFPIYAICLGFEL+TMI+
Sbjct: 124 KEGLYFEIVKKIFNKVLERNDAGEHFPIYAICLGFELLTMII 165
>ref|NP_565186.1| gamma glutamyl hydrolase, putative; protein id: At1g78680.1,
supported by cDNA: gi_3169655 [Arabidopsis thaliana]
gi|25355556|pir||F96815 hypothetical protein F9K20.28
[imported] - Arabidopsis thaliana
gi|3834325|gb|AAC83041.1| Strong similarity to
gb|AF067141 gamma-glutamyl hydrolase from Arabidopsis
thaliana. ESTs gb|R83955, gb|T45062, gb|T22220,
gb|AA586207, gb|AI099851 and gb|AI00672 come from this
gene
Length = 327
Score = 201 bits (510), Expect = 6e-51
Identities = 96/139 (69%), Positives = 116/139 (83%), Gaps = 2/139 (1%)
Frame = +1
Query: 130 ILLPNH--YNRNTPVTCPAPNPNLYYQPVIGIVTHPGDGATGRLSNATGASYIAASYVKF 303
ILLP+ ++ + C AP+PNL Y+PVIGI++HPGDGA+GRLSNAT AS IAASYVK
Sbjct: 7 ILLPSQTGFDISRSPVCSAPDPNLNYRPVIGILSHPGDGASGRLSNATDASSIAASYVKL 66
Query: 304 VEAAGARVVPLIYNEPTEKLFKKLELVNGVLFTGGWAKSGLYFETIGRIFKKALEKNDAG 483
E+ GARV+PLI+NEP E LF+KLELVNGV+ TGGWAK GLYFE + +IF K LE+NDAG
Sbjct: 67 AESGGARVIPLIFNEPEEILFQKLELVNGVILTGGWAKEGLYFEIVKKIFNKVLERNDAG 126
Query: 484 DHFPIYAICLGFELITMIV 540
+HFPIYAICLGFEL+TMI+
Sbjct: 127 EHFPIYAICLGFELLTMII 145
>ref|NP_177987.1| gamma glutamyl hydrolase, putative; protein id: At1g78660.1,
supported by cDNA: gi_16323195 [Arabidopsis thaliana]
gi|25355557|pir||D96815 probable gamma-glutamyl
hydrolase [imported] - Arabidopsis thaliana
gi|4836867|gb|AAD30570.1|AC007260_1 putative
gamma-glutamyl hydrolase [Arabidopsis thaliana]
Length = 348
Score = 198 bits (503), Expect = 4e-50
Identities = 93/147 (63%), Positives = 120/147 (81%), Gaps = 2/147 (1%)
Frame = +1
Query: 106 LSAETHPAILLPNH--YNRNTPVTCPAPNPNLYYQPVIGIVTHPGDGATGRLSNATGASY 279
L+ E +ILLP+ ++ + C +P+PNL Y+PVIGI++HPGDGA+GRL+N T ++Y
Sbjct: 20 LAKEASESILLPSESGFDGSRSPVCSSPDPNLNYRPVIGILSHPGDGASGRLTNDTSSTY 79
Query: 280 IAASYVKFVEAAGARVVPLIYNEPTEKLFKKLELVNGVLFTGGWAKSGLYFETIGRIFKK 459
IAASYVKF EA GARV+PLIYNEP E LF+KLELVNGV+FTGGWAK YFE + +IF K
Sbjct: 80 IAASYVKFAEAGGARVIPLIYNEPEEVLFQKLELVNGVIFTGGWAKKYDYFEIVKKIFTK 139
Query: 460 ALEKNDAGDHFPIYAICLGFELITMIV 540
ALE+NDAG+HFP+Y ICLGFEL+++I+
Sbjct: 140 ALERNDAGEHFPVYGICLGFELMSIII 166
>gb|AAM65646.1| gamma glutamyl hydrolase, putative [Arabidopsis thaliana]
Length = 352
Score = 176 bits (446), Expect = 2e-43
Identities = 83/130 (63%), Positives = 104/130 (79%), Gaps = 7/130 (5%)
Frame = +1
Query: 172 CPAPNPNLYYQPVIGIVTHPGDGA-TGRLSN------ATGASYIAASYVKFVEAAGARVV 330
C A +PNL Y+PVIGI+THPG+G RL + AT SYIAASYVK E GARV+
Sbjct: 47 CSAADPNLNYKPVIGILTHPGEGRWDARLHSLKNYAYATNISYIAASYVKLAETGGARVI 106
Query: 331 PLIYNEPTEKLFKKLELVNGVLFTGGWAKSGLYFETIGRIFKKALEKNDAGDHFPIYAIC 510
PLIYNEP E LF+KLELVNGV+FTGGWAK+GLY++ + +IF K +EKNDAG+HFP+YA+C
Sbjct: 107 PLIYNEPEEILFQKLELVNGVIFTGGWAKTGLYYDVVEKIFNKVMEKNDAGEHFPVYAMC 166
Query: 511 LGFELITMIV 540
LGFE+++MI+
Sbjct: 167 LGFEILSMII 176
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 529,469,948
Number of Sequences: 1393205
Number of extensions: 14079322
Number of successful extensions: 116361
Number of sequences better than 10.0: 1228
Number of HSP's better than 10.0 without gapping: 71505
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 104853
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)