Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001906A_C01 KMC001906A_c01
(552 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
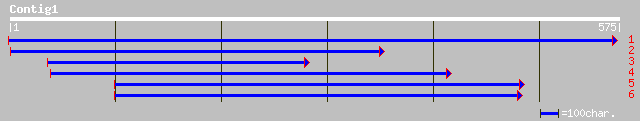
ref|NP_564969.1| Expressed protein; protein id: At1g69523.1, sup... 99 4e-20
gb|AAG60115.1|AC073178_26 hypothetical protein [Arabidopsis thal... 94 1e-18
ref|NP_564970.1| Expressed protein; protein id: At1g69526.1, sup... 91 9e-18
gb|AAM64790.1| unknown [Arabidopsis thaliana] 91 9e-18
ref|NP_564968.1| hypothetical protein; protein id: At1g69520.1 [... 86 4e-16
>ref|NP_564969.1| Expressed protein; protein id: At1g69523.1, supported by cDNA:
gi_15450408, supported by cDNA: gi_20466110 [Arabidopsis
thaliana] gi|15450409|gb|AAK96498.1| At1g69520/F10D13_17
[Arabidopsis thaliana] gi|20466111|gb|AAM19977.1|
At1g69520/F10D13_17 [Arabidopsis thaliana]
Length = 300
Score = 98.6 bits (244), Expect = 4e-20
Identities = 44/80 (55%), Positives = 61/80 (76%), Gaps = 1/80 (1%)
Frame = -3
Query: 550 LRPGGLYVFVEHVAEKDGTFL*FLQRVLDPLQQTVADGCHLSRETGNNISSAGFS-NVEL 374
LRPGG+Y+F+EHVA +DGTFL +Q VLDPLQQ VADGCHL+R TG +I A F+ ++
Sbjct: 220 LRPGGIYIFIEHVAAEDGTFLRLVQTVLDPLQQVVADGCHLTRHTGESILEARFNGGADV 279
Query: 373 EMAYLSNASFVNPHIYGIAY 314
+ LS ++++ H+YG+AY
Sbjct: 280 KKTSLSRLAYISSHVYGVAY 299
>gb|AAG60115.1|AC073178_26 hypothetical protein [Arabidopsis thaliana]
Length = 795
Score = 93.6 bits (231), Expect = 1e-18
Identities = 42/78 (53%), Positives = 59/78 (74%), Gaps = 1/78 (1%)
Frame = -3
Query: 550 LRPGGLYVFVEHVAEKDGTFL*FLQRVLDPLQQTVADGCHLSRETGNNISSAGFS-NVEL 374
LRPGG+Y+F+EHVA +DGTFL +Q VLDPLQQ VADGCHL+R TG +I A F+ ++
Sbjct: 412 LRPGGIYIFIEHVAAEDGTFLRLVQTVLDPLQQVVADGCHLTRHTGESILEARFNGGADV 471
Query: 373 EMAYLSNASFVNPHIYGI 320
+ LS ++++ H+YG+
Sbjct: 472 KKTSLSRLAYISSHVYGM 489
Score = 73.6 bits (179), Expect = 2e-12
Identities = 34/55 (61%), Positives = 43/55 (77%)
Frame = -3
Query: 550 LRPGGLYVFVEHVAEKDGTFL*FLQRVLDPLQQTVADGCHLSRETGNNISSAGFS 386
LR GG+++F+EHVA KDG+F LQ++LDPLQQ +ADGCHL+R T I AGFS
Sbjct: 181 LRQGGVFIFLEHVAAKDGSFFKRLQKLLDPLQQRLADGCHLARNTRECILEAGFS 235
Score = 60.5 bits (145), Expect = 1e-08
Identities = 31/59 (52%), Positives = 40/59 (67%), Gaps = 2/59 (3%)
Frame = -3
Query: 481 LQRVLDPLQQTVADGCHLSRETGNNISSAGF-SNVEL-EMAYLSNASFVNPHIYGIAYK 311
+Q VLDP+QQ VADGCHL+R T +IS+AGF E+ + A S + PH+YG AYK
Sbjct: 737 VQNVLDPIQQVVADGCHLTRNTDLHISAAGFDGGTEINDTAIYSFPWIIRPHVYGAAYK 795
>ref|NP_564970.1| Expressed protein; protein id: At1g69526.1, supported by cDNA:
33120. [Arabidopsis thaliana]
Length = 307
Score = 90.9 bits (224), Expect = 9e-18
Identities = 44/82 (53%), Positives = 59/82 (71%), Gaps = 2/82 (2%)
Frame = -3
Query: 550 LRPGGLYVFVEHVAEKDGTFL*FLQRVLDPLQQTVADGCHLSRETGNNISSAGF-SNVEL 374
L+PGG+++F+EHVA KDG+F +Q VLDP+QQ VADGCHL+R T +IS+AGF E+
Sbjct: 226 LKPGGIFLFIEHVAAKDGSFFRHVQNVLDPIQQVVADGCHLTRNTDLHISAAGFDGGTEI 285
Query: 373 -EMAYLSNASFVNPHIYGIAYK 311
+ A S + PH+YG AYK
Sbjct: 286 NDTAIYSFPWIIRPHVYGAAYK 307
>gb|AAM64790.1| unknown [Arabidopsis thaliana]
Length = 304
Score = 90.9 bits (224), Expect = 9e-18
Identities = 44/82 (53%), Positives = 59/82 (71%), Gaps = 2/82 (2%)
Frame = -3
Query: 550 LRPGGLYVFVEHVAEKDGTFL*FLQRVLDPLQQTVADGCHLSRETGNNISSAGF-SNVEL 374
L+PGG+++F+EHVA KDG+F +Q VLDP+QQ VADGCHL+R T +IS+AGF E+
Sbjct: 223 LKPGGIFLFIEHVAAKDGSFFRHVQNVLDPIQQVVADGCHLTRNTDLHISAAGFDGGTEI 282
Query: 373 -EMAYLSNASFVNPHIYGIAYK 311
+ A S + PH+YG AYK
Sbjct: 283 NDTAIYSFPWIIRPHVYGAAYK 304
>ref|NP_564968.1| hypothetical protein; protein id: At1g69520.1 [Arabidopsis
thaliana]
Length = 262
Score = 85.5 bits (210), Expect = 4e-16
Identities = 45/82 (54%), Positives = 57/82 (68%), Gaps = 3/82 (3%)
Frame = -3
Query: 550 LRPGGLYVFVEHVAEKDGTFL*FLQRVLDPLQQTVADGCHLSRETGNNISSAGFSNVELE 371
LR GG+++F+EHVA KDG+F LQ++LDPLQQ +ADGCHL+R T I AGFS +E
Sbjct: 181 LRQGGVFIFLEHVAAKDGSFFKRLQKLLDPLQQRLADGCHLARNTRECILEAGFSG-GVE 239
Query: 370 MAYLSNASF---VNPHIYGIAY 314
+ S SF PHIYG+AY
Sbjct: 240 VQTFSMYSFPWMTRPHIYGLAY 261
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,879,265
Number of Sequences: 1393205
Number of extensions: 10590598
Number of successful extensions: 27575
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 25354
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26852
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)