Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001903A_C01 KMC001903A_c01
(626 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
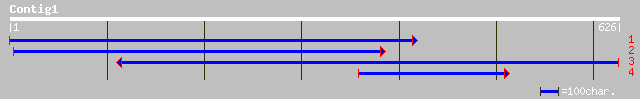
ref|NP_175036.1| histidine decarboxylase, putative; protein id: ... 154 1e-36
dbj|BAA78331.1| serine decarboxylase [Brassica napus] 149 3e-35
gb|AAG12476.2|AC037197_1 Putative histidine decarboxylase [Oryz... 57 4e-15
sp|P54772|DCHS_LYCES Histidine decarboxylase (HDC) (TOM92) gi|48... 55 3e-12
gb|ZP_00106716.1| hypothetical protein [Nostoc punctiforme] 37 1e-06
>ref|NP_175036.1| histidine decarboxylase, putative; protein id: At1g43710.1,
supported by cDNA: gi_13430641, supported by cDNA:
gi_15011301, supported by cDNA: gi_17907847 [Arabidopsis
thaliana] gi|25290978|pir||E96500 probable histidine
decarboxylase [imported] - Arabidopsis thaliana
gi|7523682|gb|AAF63121.1|AC009526_6 Putative histidine
decarboxylase [Arabidopsis thaliana]
gi|13430642|gb|AAK25943.1|AF360233_1 putative histidine
decarboxylase [Arabidopsis thaliana]
gi|14532818|gb|AAK64091.1| putative histidine
decarboxylase [Arabidopsis thaliana]
gi|15011302|gb|AAK77493.1|AF389349_1 serine
decarboxylase [Arabidopsis thaliana]
gi|17907848|dbj|BAB79456.1| histidine decarboxylase
[Arabidopsis thaliana] gi|17907853|dbj|BAB79457.1|
histidine decarboxylase [Arabidopsis thaliana]
Length = 482
Score = 154 bits (388), Expect = 1e-36
Identities = 90/159 (56%), Positives = 110/159 (68%), Gaps = 8/159 (5%)
Frame = +2
Query: 53 MVGSFGVLADDLSINGA-----VEPLPEDFDTTAIIKDPVPPVVGENGVVNGEAQVSKGK 217
MVGS L D +++ A ++ L +DFD TA++ +P+PP V NG+ A G
Sbjct: 1 MVGS---LESDQTLSMATLIEKLDILSDDFDPTAVVTEPLPPPV-TNGI---GADKGGGG 53
Query: 218 EKREIVLGRNIHTTCLEVTEPEADDEVTGDREANMASVLARYKKSLTERTKHHLGYPYNL 397
+RE+VLGRNIHTT L VTEPE +DE TGD+EA MASVLARY+K+L ERTK+HLGYPYNL
Sbjct: 54 GEREMVLGRNIHTTSLAVTEPEVNDEFTGDKEAYMASVLARYRKTLVERTKNHLGYPYNL 113
Query: 398 DFDYGALSQLQHFSINEPREIHLFESNYGC---PFPVSL 505
DFDYGAL QLQHFSIN + ESNYG PF V +
Sbjct: 114 DFDYGALGQLQHFSINNLGD-PFIESNYGVHSRPFEVGV 151
Score = 58.9 bits (141), Expect = 5e-08
Identities = 29/54 (53%), Positives = 37/54 (67%), Gaps = 1/54 (1%)
Frame = +1
Query: 457 DPFIRKQLWLSIPGQFEVGVLDWFARLWELEKNEYWGYITTVVQKAS-PGILLG 615
DPFI + FEVGVLDWFARLWE+E+++YWGYIT + + GIL+G
Sbjct: 133 DPFIESNYGVH-SRPFEVGVLDWFARLWEIERDDYWGYITNCGTEGNLHGILVG 185
>dbj|BAA78331.1| serine decarboxylase [Brassica napus]
Length = 490
Score = 149 bits (376), Expect = 3e-35
Identities = 87/158 (55%), Positives = 103/158 (65%), Gaps = 12/158 (7%)
Frame = +2
Query: 68 GVLADDLSINGA--VEPLPEDFDTTAIIKDPVP-PVVGENGVVNGEAQVSK------GKE 220
G L D S A + L E FD TA+ +P+P PV G E + K G
Sbjct: 3 GALESDQSFAMAEKFDILSEGFDPTAVAPEPLPLPVTNGTGADQEEDNLKKTKVVTNGGG 62
Query: 221 KREIVLGRNIHTTCLEVTEPEADDEVTGDREANMASVLARYKKSLTERTKHHLGYPYNLD 400
+RE+VLGRN+HTT L VTEPE++DE TGD+EA MASVLARY+K+L ERTK+HLGYPYNLD
Sbjct: 63 EREMVLGRNVHTTSLAVTEPESNDEFTGDKEAYMASVLARYRKTLVERTKYHLGYPYNLD 122
Query: 401 FDYGALSQLQHFSINEPREIHLFESNYGC---PFPVSL 505
FDYGAL QLQHFSIN + ESNYG PF V +
Sbjct: 123 FDYGALGQLQHFSINNLGD-PFIESNYGVHSRPFEVGV 159
Score = 58.9 bits (141), Expect = 5e-08
Identities = 29/54 (53%), Positives = 37/54 (67%), Gaps = 1/54 (1%)
Frame = +1
Query: 457 DPFIRKQLWLSIPGQFEVGVLDWFARLWELEKNEYWGYITTVVQKAS-PGILLG 615
DPFI + FEVGVLDWFARLWE+E+++YWGYIT + + GIL+G
Sbjct: 141 DPFIESNYGVH-SRPFEVGVLDWFARLWEIERDDYWGYITNCGTEGNLHGILVG 193
>gb|AAG12476.2|AC037197_1 Putative histidine decarboxylase [Oryza sativa]
gi|18542940|gb|AAL75763.1|AC069324_20 Putative histidine
decarboxylase [Oryza sativa]
Length = 467
Score = 56.6 bits (135), Expect(2) = 4e-15
Identities = 23/61 (37%), Positives = 40/61 (64%)
Frame = +2
Query: 263 LEVTEPEADDEVTGDREANMASVLARYKKSLTERTKHHLGYPYNLDFDYGALSQLQHFSI 442
LE+ EP AD+ +++A ++ ++A Y + L R+ +HLGYP N D+D+ L+ +FS+
Sbjct: 46 LEIEEPPADEMEAAEKKAGISRLMAGYVQHLQHRSAYHLGYPLNFDYDFSPLAPFLNFSL 105
Query: 443 N 445
N
Sbjct: 106 N 106
Score = 46.2 bits (108), Expect(2) = 4e-15
Identities = 15/27 (55%), Positives = 24/27 (88%)
Frame = +1
Query: 499 QFEVGVLDWFARLWELEKNEYWGYITT 579
QFEV VL+WFA W+++++++WGYIT+
Sbjct: 123 QFEVAVLNWFANFWDVQRDQFWGYITS 149
>sp|P54772|DCHS_LYCES Histidine decarboxylase (HDC) (TOM92) gi|481829|pir||S39554
histidine decarboxylase (EC 4.1.1.22) - tomato
gi|416534|emb|CAA50719.1| histidine decarboxylase
[Lycopersicon esculentum]
Length = 413
Score = 55.5 bits (132), Expect(2) = 3e-12
Identities = 26/48 (54%), Positives = 31/48 (64%)
Frame = +1
Query: 436 FHKRT*RDPFIRKQLWLSIPGQFEVGVLDWFARLWELEKNEYWGYITT 579
FH DPF + FEV VLDWFA+LWE+EK+EYWGYIT+
Sbjct: 53 FHLNNCGDPFTQHPTDFHSK-DFEVAVLDWFAQLWEIEKDEYWGYITS 99
Score = 37.7 bits (86), Expect(2) = 3e-12
Identities = 16/40 (40%), Positives = 29/40 (72%), Gaps = 1/40 (2%)
Frame = +2
Query: 329 VLARYKKSLTERTKHHLGYPYNLDFDYGA-LSQLQHFSIN 445
+L +Y ++L+ER K+H+GYP N+ +++ A L+ L F +N
Sbjct: 17 ILTQYLETLSERKKYHIGYPINMCYEHHATLAPLLQFHLN 56
>gb|ZP_00106716.1| hypothetical protein [Nostoc punctiforme]
Length = 384
Score = 37.4 bits (85), Expect(2) = 1e-06
Identities = 17/57 (29%), Positives = 33/57 (57%)
Frame = +2
Query: 314 ANMASVLARYKKSLTERTKHHLGYPYNLDFDYGALSQLQHFSINEPREIHLFESNYG 484
+ +A LA + + +R++ H GYPYNL DY A+ + + +N + ++ E ++G
Sbjct: 3 SKVAKELADFLLQIEQRSQFHAGYPYNLSCDYSAIGKFFNHLLNNAGDPYI-EPDFG 58
Score = 37.0 bits (84), Expect(2) = 1e-06
Identities = 16/40 (40%), Positives = 26/40 (65%)
Frame = +1
Query: 457 DPFIRKQLWLSIPGQFEVGVLDWFARLWELEKNEYWGYIT 576
DP+I L +FE VL +FA L+++ +N++WGY+T
Sbjct: 50 DPYIEPDFGLH-SRKFEQEVLSFFAHLYKIPENQFWGYVT 88
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 598,792,144
Number of Sequences: 1393205
Number of extensions: 13790731
Number of successful extensions: 38381
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 36467
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38316
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25586195130
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)