Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001897A_C01 KMC001897A_c01
(929 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
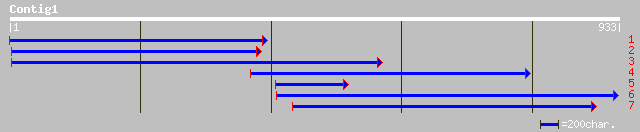
gb|AAM61665.1| leucoanthocyanidin dioxygenase-like protein [Arab... 187 3e-46
ref|NP_196179.1| leucoanthocyanidin dioxygenase-like protein; pr... 187 3e-46
gb|AAD52015.1|AF082862_1 unknown [Pisum sativum] 186 4e-46
ref|NP_187728.1| putative leucoanthocyanidin dioxygenase; protei... 181 1e-44
ref|NP_191156.1| leucoanthocyanidin dioxygenase -like protein; p... 175 1e-42
>gb|AAM61665.1| leucoanthocyanidin dioxygenase-like protein [Arabidopsis thaliana]
Length = 355
Score = 187 bits (474), Expect = 3e-46
Identities = 92/119 (77%), Positives = 100/119 (83%)
Frame = -2
Query: 928 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 749
LLPDDQV GLQVRK D WITVKP PHAFIVNIGDQIQ+LSN+ YKSVEHRVIVNS+KERV
Sbjct: 237 LLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERV 296
Query: 748 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 572
SLAFFYNP+ DIPI+P +ELV P LY MTFD+YR FIR +GP GKSHVES SPR
Sbjct: 297 SLAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQGPQGKSHVESHISPR 355
>ref|NP_196179.1| leucoanthocyanidin dioxygenase-like protein; protein id:
At5g05600.1, supported by cDNA: 13012., supported by
cDNA: gi_14532537 [Arabidopsis thaliana]
gi|10178137|dbj|BAB11549.1| leucoanthocyanidin
dioxygenase-like protein [Arabidopsis thaliana]
gi|14532538|gb|AAK63997.1| AT5g05600/MOP10_14
[Arabidopsis thaliana] gi|22137300|gb|AAM91495.1|
AT5g05600/MOP10_14 [Arabidopsis thaliana]
Length = 371
Score = 187 bits (474), Expect = 3e-46
Identities = 92/119 (77%), Positives = 100/119 (83%)
Frame = -2
Query: 928 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 749
LLPDDQV GLQVRK D WITVKP PHAFIVNIGDQIQ+LSN+ YKSVEHRVIVNS+KERV
Sbjct: 253 LLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERV 312
Query: 748 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 572
SLAFFYNP+ DIPI+P +ELV P LY MTFD+YR FIR +GP GKSHVES SPR
Sbjct: 313 SLAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQGPQGKSHVESHISPR 371
>gb|AAD52015.1|AF082862_1 unknown [Pisum sativum]
Length = 134
Score = 186 bits (472), Expect = 4e-46
Identities = 88/117 (75%), Positives = 105/117 (89%)
Frame = -2
Query: 928 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 749
LLPDD V+GLQVRKG++WITVKPVP+AFI+NIGDQIQV+SNAIYKSVEHRVIVN ++RV
Sbjct: 16 LLPDDFVSGLQVRKGNDWITVKPVPNAFIINIGDQIQVMSNAIYKSVEHRVIVNPTQDRV 75
Query: 748 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKS 578
SLA FYNP+ D+ I+PAKELV +++PALY MT+DEYR +IRMKGPCGK+ VESL S
Sbjct: 76 SLAMFYNPKSDLIIQPAKELVTKERPALYPPMTYDEYRLYIRMKGPCGKAQVESLAS 132
>ref|NP_187728.1| putative leucoanthocyanidin dioxygenase; protein id: At3g11180.1
[Arabidopsis thaliana]
gi|6016680|gb|AAF01507.1|AC009991_3 putative
leucoanthocyanidin dioxygenase [Arabidopsis thaliana]
gi|12321884|gb|AAG50980.1|AC073395_22 leucoanthocyanidin
dioxygenase, putative; 41415-43854 [Arabidopsis
thaliana]
Length = 400
Score = 181 bits (460), Expect = 1e-44
Identities = 89/119 (74%), Positives = 98/119 (81%)
Frame = -2
Query: 928 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 749
LLPDDQV GLQVR GD WITV P+ HAFIVNIGDQIQ+LSN+ YKSVEHRVIVNS KERV
Sbjct: 282 LLPDDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEHRVIVNSEKERV 341
Query: 748 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 572
SLAFFYNP+ DIPI+P ++LV P LY MTFD+YR FIR +GP GKSHVES SPR
Sbjct: 342 SLAFFYNPKSDIPIQPMQQLVTSTMPPLYPPMTFDQYRLFIRTQGPRGKSHVESHISPR 400
>ref|NP_191156.1| leucoanthocyanidin dioxygenase -like protein; protein id:
At3g55970.1 [Arabidopsis thaliana]
gi|11255840|pir||T49209 leucoanthocyanidin
dioxygenase-like protein - Arabidopsis thaliana
gi|7573492|emb|CAB87851.1| leucoanthocyanidin
dioxygenase-like protein [Arabidopsis thaliana]
gi|12043537|emb|CAC19787.1| putative leucoanthocyanidin
dioxygenase [Arabidopsis thaliana]
Length = 363
Score = 175 bits (443), Expect = 1e-42
Identities = 83/120 (69%), Positives = 100/120 (83%), Gaps = 1/120 (0%)
Frame = -2
Query: 928 LLPDDQVTGLQVRKGDN-WITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKER 752
LLPD+QV LQVR D+ WITV+P PHAFIVN+GDQIQ+LSN+IYKSVEHRVIVN ER
Sbjct: 244 LLPDEQVASLQVRGSDDAWITVEPAPHAFIVNMGDQIQMLSNSIYKSVEHRVIVNPENER 303
Query: 751 VSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 572
+SLAFFYNP+G++PIEP KELV D PALY++ T+D YR+FIR +GP K H++ LKSPR
Sbjct: 304 LSLAFFYNPKGNVPIEPLKELVTVDSPALYSSTTYDRYRQFIRTQGPRSKCHIDELKSPR 363
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 823,493,601
Number of Sequences: 1393205
Number of extensions: 18733144
Number of successful extensions: 40704
Number of sequences better than 10.0: 635
Number of HSP's better than 10.0 without gapping: 39181
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40610
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51582455952
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)