Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001864A_C01 KMC001864A_c01
(609 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
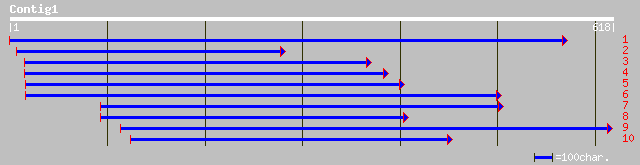
ref|NP_726773.2| trithorax-related CG3848-PD gi|28381556|gb|AAN0... 37 0.15
gb|AAM29656.1| SD13650p [Drosophila melanogaster] 37 0.15
pir||T12687 ALR protein homolog - fruit fly (Drosophila melanoga... 37 0.15
ref|NP_525040.2| trithorax-related CG3848-PC gi|22831528|gb|AAF4... 37 0.15
ref|NP_472935.1| rifin [Plasmodium falciparum 3D7] gi|7494409|pi... 35 1.00
>ref|NP_726773.2| trithorax-related CG3848-PD gi|28381556|gb|AAN09063.2| CG3848-PD
[Drosophila melanogaster]
Length = 2431
Score = 37.4 bits (85), Expect = 0.15
Identities = 27/86 (31%), Positives = 38/86 (43%), Gaps = 15/86 (17%)
Frame = -1
Query: 603 QGIGSKAPRGSLVDHPGFYSLPSTEAAGVSV---PEEE---------RPHNLPSQR---A 469
Q G A + S D PG + ST A + V PE+E P N+P+QR
Sbjct: 1787 QPAGGSAVKSSNGDSPGSFCASSTAPAEMVVKQEPEDEDEKTPSVPGNPTNIPAQRKCIV 1846
Query: 468 DCSTESCLTSHESSGGLVLEGSPSEG 391
C + C T+ + GL L+G+ G
Sbjct: 1847 KCFSADCFTTDSAPSGLELDGTAGAG 1872
>gb|AAM29656.1| SD13650p [Drosophila melanogaster]
Length = 1019
Score = 37.4 bits (85), Expect = 0.15
Identities = 27/86 (31%), Positives = 38/86 (43%), Gaps = 15/86 (17%)
Frame = -1
Query: 603 QGIGSKAPRGSLVDHPGFYSLPSTEAAGVSV---PEEE---------RPHNLPSQR---A 469
Q G A + S D PG + ST A + V PE+E P N+P+QR
Sbjct: 375 QPAGGSAVKSSNGDSPGSFCASSTAPAEMVVKQEPEDEDEKTPSVPGNPTNIPAQRKCIV 434
Query: 468 DCSTESCLTSHESSGGLVLEGSPSEG 391
C + C T+ + GL L+G+ G
Sbjct: 435 KCFSADCFTTDSAPSGLELDGTAGAG 460
>pir||T12687 ALR protein homolog - fruit fly (Drosophila melanogaster)
gi|3256105|emb|CAA15944.1|
/prediction=(method:''genscan'',
version:''1.0'')~/prediction=(method:''genefinder'',
version:''084'')~/match=(desc:''ZINC FINGER PROTEIN HRX
(ALL-1)'', species:''HOMO SAPIENS (HUMAN)'',
ranges:(query:22223..22333,
target:SWISS-PROT::Q03164:1903..1867, score:''75.00''),
(query:21936..22169,
target:SWISS-PROT::Q03164:1977..1900, score:''185.00''),
(query:21236..21409,
target:SWISS-PROT::Q03164:3747..3690, score:''67.00''),
(query:20787..20918,
target:SWISS-PROT::Q03164:3878..3835, score:''106.00''),
(query>
Length = 2422
Score = 37.4 bits (85), Expect = 0.15
Identities = 27/86 (31%), Positives = 38/86 (43%), Gaps = 15/86 (17%)
Frame = -1
Query: 603 QGIGSKAPRGSLVDHPGFYSLPSTEAAGVSV---PEEE---------RPHNLPSQR---A 469
Q G A + S D PG + ST A + V PE+E P N+P+QR
Sbjct: 1778 QPAGGSAVKSSNGDSPGSFCASSTAPAEMVVKQEPEDEDEKTPSVPGNPTNIPAQRKCIV 1837
Query: 468 DCSTESCLTSHESSGGLVLEGSPSEG 391
C + C T+ + GL L+G+ G
Sbjct: 1838 KCFSADCFTTDSAPSGLELDGTAGAG 1863
>ref|NP_525040.2| trithorax-related CG3848-PC gi|22831528|gb|AAF45684.2| CG3848-PC
[Drosophila melanogaster]
Length = 2410
Score = 37.4 bits (85), Expect = 0.15
Identities = 27/86 (31%), Positives = 38/86 (43%), Gaps = 15/86 (17%)
Frame = -1
Query: 603 QGIGSKAPRGSLVDHPGFYSLPSTEAAGVSV---PEEE---------RPHNLPSQR---A 469
Q G A + S D PG + ST A + V PE+E P N+P+QR
Sbjct: 1766 QPAGGSAVKSSNGDSPGSFCASSTAPAEMVVKQEPEDEDEKTPSVPGNPTNIPAQRKCIV 1825
Query: 468 DCSTESCLTSHESSGGLVLEGSPSEG 391
C + C T+ + GL L+G+ G
Sbjct: 1826 KCFSADCFTTDSAPSGLELDGTAGAG 1851
>ref|NP_472935.1| rifin [Plasmodium falciparum 3D7] gi|7494409|pir||H71624 rifin
PFB0030c - malaria parasite (Plasmodium falciparum)
gi|3845076|gb|AAC71796.1| rifin [Plasmodium falciparum
3D7]
Length = 370
Score = 34.7 bits (78), Expect = 1.00
Identities = 32/109 (29%), Positives = 51/109 (46%), Gaps = 10/109 (9%)
Frame = -1
Query: 471 ADCSTESCLTSHESSGGLVLEGSPSEGRRRMLGMDSF-AAPLIWSE--AKMKTQAINLAH 301
AD + + CL GG VL+ S G LG+D++ AA L+ ++ A+ A LA
Sbjct: 131 ADKTEKFCLNCGVQLGGGVLQASGLLGGIGQLGLDAWKAAALVTAKELAEKAGAAKGLAE 190
Query: 300 GNSHGISRYGM*GLMVILKLMVHSL-------IIRGTIELLSFSYRYLN 175
GN+HG +K+++H L ++ G E +S + Y N
Sbjct: 191 GNAHG------------MKIVIHHLKELHIDKLVPGICEKISSTGHYAN 227
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 529,675,682
Number of Sequences: 1393205
Number of extensions: 11573756
Number of successful extensions: 24079
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 23233
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24065
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24283162270
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)