Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001862A_C01 KMC001862A_c01
(544 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
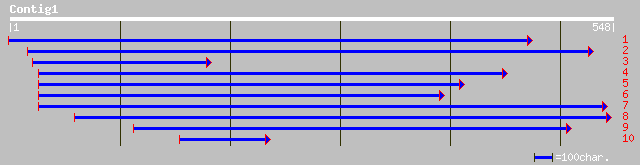
gb|AAK43508.1|AC020666_18 putative WW-domain oxidoreductase [Ory... 92 4e-18
ref|NP_192880.1| putative protein; protein id: At4g11410.1 [Arab... 88 6e-17
ref|NP_567681.1| putative protein; protein id: At4g23430.1, supp... 84 1e-15
gb|AAN13078.1| unknown protein [Arabidopsis thaliana] 83 2e-15
ref|NP_194073.1| putative protein; protein id: At4g23420.1 [Arab... 83 2e-15
>gb|AAK43508.1|AC020666_18 putative WW-domain oxidoreductase [Oryza sativa (japonica
cultivar-group)]
Length = 319
Score = 92.0 bits (227), Expect = 4e-18
Identities = 38/66 (57%), Positives = 54/66 (81%)
Frame = -1
Query: 541 IINVVGRMVMKNVQQGAATTCYVALHPEVKGVSGKYFSDSNVSKTTPHGTDADLAKKLWD 362
++N +GR+V K V+QGAATTCYVALHP+V G+SGKYFS+ N+ + ++A+LAKKLW+
Sbjct: 249 VVNTIGRIVCKTVEQGAATTCYVALHPQVTGISGKYFSNCNLETPSSQASNAELAKKLWE 308
Query: 361 FSMNLI 344
FS N++
Sbjct: 309 FSSNIV 314
>ref|NP_192880.1| putative protein; protein id: At4g11410.1 [Arabidopsis thaliana]
gi|7486117|pir||T10561 hypothetical protein F25E4.30 -
Arabidopsis thaliana gi|7267840|emb|CAB81242.1| putative
protein [Arabidopsis thaliana]
gi|7321038|emb|CAB82146.1| putative protein [Arabidopsis
thaliana]
Length = 317
Score = 88.2 bits (217), Expect = 6e-17
Identities = 38/63 (60%), Positives = 48/63 (75%)
Frame = -1
Query: 535 NVVGRMVMKNVQQGAATTCYVALHPEVKGVSGKYFSDSNVSKTTPHGTDADLAKKLWDFS 356
N VG+ V+K++ QGAATTCY ALHP+ KGVSG+Y D+N+S G D DLAKKLW+FS
Sbjct: 249 NAVGKYVLKSIPQGAATTCYAALHPQAKGVSGEYLMDNNISDPNSQGKDKDLAKKLWEFS 308
Query: 355 MNL 347
+ L
Sbjct: 309 LRL 311
>ref|NP_567681.1| putative protein; protein id: At4g23430.1, supported by cDNA: 4578.
[Arabidopsis thaliana] gi|21593805|gb|AAM65772.1|
putativepod-specific dehydrogenase SAC25 [Arabidopsis
thaliana]
Length = 320
Score = 84.0 bits (206), Expect = 1e-15
Identities = 37/64 (57%), Positives = 49/64 (75%)
Frame = -1
Query: 538 INVVGRMVMKNVQQGAATTCYVALHPEVKGVSGKYFSDSNVSKTTPHGTDADLAKKLWDF 359
+ V + ++K+V QGAATTCYVAL+P+V GVSG+YF DSN++K P D +LAKK+WDF
Sbjct: 247 VGAVAKYILKSVPQGAATTCYVALNPQVAGVSGEYFQDSNIAKPLPLVKDTELAKKVWDF 306
Query: 358 SMNL 347
S L
Sbjct: 307 STKL 310
>gb|AAN13078.1| unknown protein [Arabidopsis thaliana]
Length = 316
Score = 83.2 bits (204), Expect = 2e-15
Identities = 38/66 (57%), Positives = 50/66 (75%)
Frame = -1
Query: 544 GIINVVGRMVMKNVQQGAATTCYVALHPEVKGVSGKYFSDSNVSKTTPHGTDADLAKKLW 365
G + V + ++K+V QGAATTCYVAL+P+V GV+G+YFSDSN++K D +LAKKLW
Sbjct: 247 GAVGAVAKYMVKSVPQGAATTCYVALNPQVAGVTGEYFSDSNIAKPIELVKDTELAKKLW 306
Query: 364 DFSMNL 347
DFS L
Sbjct: 307 DFSTKL 312
>ref|NP_194073.1| putative protein; protein id: At4g23420.1 [Arabidopsis thaliana]
gi|7485550|pir||T05380 hypothetical protein F16G20.120 -
Arabidopsis thaliana gi|3451067|emb|CAA20463.1| putative
protein [Arabidopsis thaliana]
gi|7269190|emb|CAB79297.1| putative protein [Arabidopsis
thaliana]
Length = 175
Score = 83.2 bits (204), Expect = 2e-15
Identities = 38/66 (57%), Positives = 50/66 (75%)
Frame = -1
Query: 544 GIINVVGRMVMKNVQQGAATTCYVALHPEVKGVSGKYFSDSNVSKTTPHGTDADLAKKLW 365
G + V + ++K+V QGAATTCYVAL+P+V GV+G+YFSDSN++K D +LAKKLW
Sbjct: 106 GAVGAVAKYMVKSVPQGAATTCYVALNPQVAGVTGEYFSDSNIAKPIELVKDTELAKKLW 165
Query: 364 DFSMNL 347
DFS L
Sbjct: 166 DFSTKL 171
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,302,841
Number of Sequences: 1393205
Number of extensions: 10063314
Number of successful extensions: 19317
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 18839
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19311
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18750593680
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)