Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001856A_C01 KMC001856A_c01
(437 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
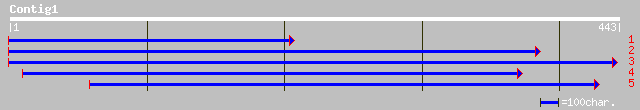
pir||T52337 phosphoprotein phosphatase (EC 3.1.3.16) 2C [importe... 81 3e-15
ref|NP_191785.1| protein phosphatase 2C (PP2C); protein id: At3g... 72 2e-12
dbj|BAB12036.1| putative protein phosphatase [Oryza sativa (japo... 64 4e-10
ref|NP_188351.1| putative protein phosphatase; protein id: At3g1... 51 4e-06
dbj|BAB02728.1| protein phosphatase 2C-like protein [Arabidopsis... 51 4e-06
>pir||T52337 phosphoprotein phosphatase (EC 3.1.3.16) 2C [imported] - common ice
plant gi|3643085|gb|AAC36698.1| protein phosphatase-2C;
PP2C [Mesembryanthemum crystallinum]
Length = 359
Score = 81.3 bits (199), Expect = 3e-15
Identities = 43/64 (67%), Positives = 50/64 (77%)
Frame = -1
Query: 437 ARDLVMEALRLNTFDNLTVIIVCFSSLDHRESEPSPPQRKLRCCSLSAEALCSLRSLLEG 258
A+DLV EALR +T DNLTVIIVCFSS+DH+ + P R+ R CSLSAEAL SLRSLLEG
Sbjct: 297 AKDLVNEALRRHTIDNLTVIIVCFSSIDHQREQTGPRPRRFR-CSLSAEALSSLRSLLEG 355
Query: 257 GASN 246
S+
Sbjct: 356 NESH 359
>ref|NP_191785.1| protein phosphatase 2C (PP2C); protein id: At3g62260.1, supported
by cDNA: 20050. [Arabidopsis thaliana]
gi|11358069|pir||T48018 hypothetical protein T17J13.220
- Arabidopsis thaliana gi|6899936|emb|CAB71886.1|
putative protein [Arabidopsis thaliana]
gi|21554078|gb|AAM63159.1| protein phosphatase-2C
[Arabidopsis thaliana]
Length = 383
Score = 71.6 bits (174), Expect = 2e-12
Identities = 36/60 (60%), Positives = 45/60 (75%)
Frame = -1
Query: 437 ARDLVMEALRLNTFDNLTVIIVCFSSLDHRESEPSPPQRKLRCCSLSAEALCSLRSLLEG 258
AR+LVMEAL N+FDNLT ++VCF ++D R +P P K RC SLS EA CSLR+LL+G
Sbjct: 325 ARELVMEALGRNSFDNLTAVVVCFMTMD-RGDKPVVPLEKRRCFSLSPEAFCSLRNLLDG 383
>dbj|BAB12036.1| putative protein phosphatase [Oryza sativa (japonica
cultivar-group)]
Length = 380
Score = 63.9 bits (154), Expect = 4e-10
Identities = 35/62 (56%), Positives = 43/62 (68%), Gaps = 3/62 (4%)
Frame = -1
Query: 437 ARDLVMEALRLNTFDNLTVIIVCFSSL--DHRESEPSPPQRKLRCC-SLSAEALCSLRSL 267
AR+L MEA RL TFDNLTVI++CF S S P R++RCC SLS+EALC+L+
Sbjct: 316 ARELAMEAKRLQTFDNLTVIVICFGSELGGGSPSSEQAPIRRVRCCKSLSSEALCNLKKW 375
Query: 266 LE 261
LE
Sbjct: 376 LE 377
>ref|NP_188351.1| putative protein phosphatase; protein id: At3g17250.1 [Arabidopsis
thaliana]
Length = 378
Score = 50.8 bits (120), Expect = 4e-06
Identities = 27/60 (45%), Positives = 42/60 (70%)
Frame = -1
Query: 437 ARDLVMEALRLNTFDNLTVIIVCFSSLDHRESEPSPPQRKLRCCSLSAEALCSLRSLLEG 258
A +L EALRL++ DN+TV+++CFS S P+P +R++R C +S EA L+++LEG
Sbjct: 326 AMELGREALRLDSSDNVTVVVICFS------SSPAPQRRRIRFC-VSDEARARLQTMLEG 378
>dbj|BAB02728.1| protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 422
Score = 50.8 bits (120), Expect = 4e-06
Identities = 27/60 (45%), Positives = 42/60 (70%)
Frame = -1
Query: 437 ARDLVMEALRLNTFDNLTVIIVCFSSLDHRESEPSPPQRKLRCCSLSAEALCSLRSLLEG 258
A +L EALRL++ DN+TV+++CFS S P+P +R++R C +S EA L+++LEG
Sbjct: 370 AMELGREALRLDSSDNVTVVVICFS------SSPAPQRRRIRFC-VSDEARARLQTMLEG 422
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 389,775,590
Number of Sequences: 1393205
Number of extensions: 8063305
Number of successful extensions: 17283
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 16850
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17274
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 6722674608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)