Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
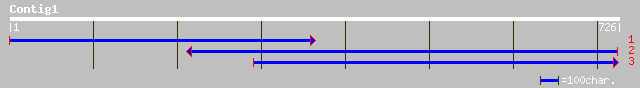
Query= KMC001839A_C01 KMC001839A_c01
(726 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA10101.1| ribosomal protein S28 [Prunus persica] gi|394772... 98 7e-22
emb|CAA10104.1| ribosomal protein S28 [Prunus persica] 96 4e-21
gb|AAM78552.1|AF522185_1 ribosomal protein small subunit 28 kDa ... 96 4e-19
ref|NP_187620.1| putative ribosomal protein S28; protein id: At3... 87 5e-19
emb|CAA04565.1| rpS28 [Hordeum vulgare subsp. vulgare] 95 9e-19
>emb|CAA10101.1| ribosomal protein S28 [Prunus persica] gi|3947721|emb|CAA10102.1|
ribosomal protein S28 [Prunus persica]
gi|3947723|emb|CAA10103.1| ribosomal protein S28 [Prunus
persica]
Length = 65
Score = 98.2 bits (243), Expect(2) = 7e-22
Identities = 50/54 (92%), Positives = 51/54 (93%)
Frame = -3
Query: 628 KLWVRTGSRGQVTQVRVKFLDDQNRHIMRNVKGPVREGDILTLLESEREARRLR 467
K+ RTGSRGQVTQVRVKFLDDQNR IMRNVKGPVREGDILTLLESEREARRLR
Sbjct: 12 KVMGRTGSRGQVTQVRVKFLDDQNRFIMRNVKGPVREGDILTLLESEREARRLR 65
Score = 28.1 bits (61), Expect(2) = 7e-22
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = -2
Query: 662 MESQVKHAIVVKIMG 618
M+S VKHA+VVK+MG
Sbjct: 1 MDSTVKHAVVVKVMG 15
>emb|CAA10104.1| ribosomal protein S28 [Prunus persica]
Length = 65
Score = 95.5 bits (236), Expect(2) = 4e-21
Identities = 49/54 (90%), Positives = 50/54 (91%)
Frame = -3
Query: 628 KLWVRTGSRGQVTQVRVKFLDDQNRHIMRNVKGPVREGDILTLLESEREARRLR 467
K+ RTGSRGQVTQVRVKFL DQNR IMRNVKGPVREGDILTLLESEREARRLR
Sbjct: 12 KVMGRTGSRGQVTQVRVKFLGDQNRFIMRNVKGPVREGDILTLLESEREARRLR 65
Score = 28.1 bits (61), Expect(2) = 4e-21
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = -2
Query: 662 MESQVKHAIVVKIMG 618
M+S VKHA+VVK+MG
Sbjct: 1 MDSTVKHAVVVKVMG 15
>gb|AAM78552.1|AF522185_1 ribosomal protein small subunit 28 kDa [Helianthus annuus]
Length = 65
Score = 96.3 bits (238), Expect = 4e-19
Identities = 48/54 (88%), Positives = 51/54 (93%)
Frame = -3
Query: 628 KLWVRTGSRGQVTQVRVKFLDDQNRHIMRNVKGPVREGDILTLLESEREARRLR 467
K+ R+GSRGQVTQVRVKFLDDQNR IMRNVKGPVREGD+LTLLESEREARRLR
Sbjct: 12 KVMGRSGSRGQVTQVRVKFLDDQNRFIMRNVKGPVREGDVLTLLESEREARRLR 65
Score = 32.0 bits (71), Expect = 9.1
Identities = 16/27 (59%), Positives = 19/27 (70%)
Frame = -2
Query: 662 MESQVKHAIVVKIMGPYWIQRSGHSSQ 582
MESQ+KHA+VVK+MG RSG Q
Sbjct: 1 MESQIKHAVVVKVMG-----RSGSRGQ 22
>ref|NP_187620.1| putative ribosomal protein S28; protein id: At3g10090.1
[Arabidopsis thaliana] gi|15237548|ref|NP_196005.1|
RIBOSOMAL PROTEIN S28- like; protein id: At5g03850.1
[Arabidopsis thaliana] gi|11276649|pir||T48412 RIBOSOMAL
PROTEIN S28-like - Arabidopsis thaliana
gi|6143868|gb|AAF04415.1|AC010927_8 putative ribosomal
protein S28 [Arabidopsis thaliana]
gi|7406395|emb|CAB85505.1| RIBOSOMAL PROTEIN S28-like
[Arabidopsis thaliana] gi|9758014|dbj|BAB08611.1|
ribosomal protein S28 [Arabidopsis thaliana]
gi|22136042|gb|AAM91603.1| ribosomal protein S28-like
protein [Arabidopsis thaliana]
gi|23197756|gb|AAN15405.1| ribosomal protein S28-like
protein [Arabidopsis thaliana]
Length = 64
Score = 86.7 bits (213), Expect(2) = 5e-19
Identities = 46/54 (85%), Positives = 49/54 (90%)
Frame = -3
Query: 628 KLWVRTGSRGQVTQVRVKFLDDQNRHIMRNVKGPVREGDILTLLESEREARRLR 467
K+ RTGSRGQVTQVRVKF D +R+IMRNVKGPVREGDILTLLESEREARRLR
Sbjct: 12 KVMGRTGSRGQVTQVRVKFTDS-DRYIMRNVKGPVREGDILTLLESEREARRLR 64
Score = 30.0 bits (66), Expect(2) = 5e-19
Identities = 11/15 (73%), Positives = 15/15 (99%)
Frame = -2
Query: 662 MESQVKHAIVVKIMG 618
M+SQ+KHA+VVK+MG
Sbjct: 1 MDSQIKHAVVVKVMG 15
>emb|CAA04565.1| rpS28 [Hordeum vulgare subsp. vulgare]
Length = 65
Score = 95.1 bits (235), Expect = 9e-19
Identities = 49/59 (83%), Positives = 53/59 (89%)
Frame = -3
Query: 643 MLLLSKLWVRTGSRGQVTQVRVKFLDDQNRHIMRNVKGPVREGDILTLLESEREARRLR 467
+ ++ K+ RTGSRGQVTQVRVKFLD QNR IMRNVKGPVREGDILTLLESEREARRLR
Sbjct: 7 LAVVVKVMGRTGSRGQVTQVRVKFLDHQNRLIMRNVKGPVREGDILTLLESEREARRLR 65
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 573,600,070
Number of Sequences: 1393205
Number of extensions: 11773265
Number of successful extensions: 29823
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 28495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29769
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34062062287
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)