Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
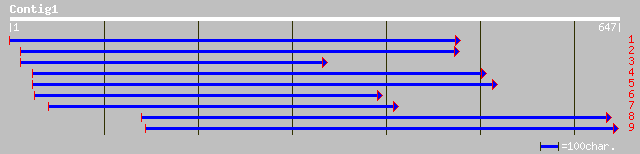
Query= KMC001819A_C01 KMC001819A_c01
(632 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL38258.1| dnaJ-like protein [Arabidopsis thaliana] gi|27311... 131 6e-33
ref|NP_191293.1| dnaJ-like protein; protein id: At3g57340.1, sup... 131 6e-33
dbj|BAB89081.1| dnaJ-like protein [Oryza sativa (japonica cultiv... 125 2e-30
ref|NP_199717.1| DNAJ-like protein; protein id: At5g49060.1 [Ara... 61 1e-08
ref|NP_060096.1| DnaJ (Hsp40) homolog, subfamily B, member 12 [H... 53 4e-06
>gb|AAL38258.1| dnaJ-like protein [Arabidopsis thaliana] gi|27311873|gb|AAO00902.1|
dnaJ-like protein [Arabidopsis thaliana]
Length = 367
Score = 131 bits (330), Expect(2) = 6e-33
Identities = 56/95 (58%), Positives = 79/95 (82%)
Frame = -2
Query: 631 FNARALIQLFPVLLILLINFLPSSEPIYTLSQSYPYEHRVTTPRGVNYYVKSTKFAQDYP 452
FNAR L+QL PV+ ILL+NF+PSS+P+Y LS +YPY+++ TT +GVNY+VKS+KF QDYP
Sbjct: 254 FNARILLQLLPVVFILLLNFMPSSQPVYQLSATYPYQYKFTTQKGVNYFVKSSKFEQDYP 313
Query: 451 EGTRERETVEDKVEREYFGILRQNCQFELQRRQCG 347
+ +R T+E++VER+Y IL QNC++E+QR+Q G
Sbjct: 314 RDSNDRHTLEEQVERDYVSILSQNCRYEMQRKQWG 348
Score = 31.2 bits (69), Expect(2) = 6e-33
Identities = 8/16 (50%), Positives = 16/16 (100%)
Frame = -3
Query: 348 GYIKETPHCDILRKYE 301
G+++ETPHCD++R+++
Sbjct: 348 GFVRETPHCDMMRRFD 363
>ref|NP_191293.1| dnaJ-like protein; protein id: At3g57340.1, supported by cDNA:
120719., supported by cDNA: gi_17473564 [Arabidopsis
thaliana] gi|11357271|pir||T45812 dnaJ-like protein -
Arabidopsis thaliana gi|6735313|emb|CAB68140.1|
dnaJ-like protein [Arabidopsis thaliana]
Length = 367
Score = 131 bits (330), Expect(2) = 6e-33
Identities = 56/95 (58%), Positives = 79/95 (82%)
Frame = -2
Query: 631 FNARALIQLFPVLLILLINFLPSSEPIYTLSQSYPYEHRVTTPRGVNYYVKSTKFAQDYP 452
FNAR L+QL PV+ ILL+NF+PSS+P+Y LS +YPY+++ TT +GVNY+VKS+KF QDYP
Sbjct: 254 FNARILLQLLPVVFILLLNFMPSSQPVYQLSATYPYQYKFTTQKGVNYFVKSSKFEQDYP 313
Query: 451 EGTRERETVEDKVEREYFGILRQNCQFELQRRQCG 347
+ +R T+E++VER+Y IL QNC++E+QR+Q G
Sbjct: 314 RDSNDRHTLEEQVERDYVSILSQNCRYEMQRKQWG 348
Score = 31.2 bits (69), Expect(2) = 6e-33
Identities = 8/16 (50%), Positives = 16/16 (100%)
Frame = -3
Query: 348 GYIKETPHCDILRKYE 301
G+++ETPHCD++R+++
Sbjct: 348 GFVRETPHCDMMRRFD 363
>dbj|BAB89081.1| dnaJ-like protein [Oryza sativa (japonica cultivar-group)]
Length = 380
Score = 125 bits (313), Expect(2) = 2e-30
Identities = 59/93 (63%), Positives = 71/93 (75%)
Frame = -2
Query: 622 RALIQLFPVLLILLINFLPSSEPIYTLSQSYPYEHRVTTPRGVNYYVKSTKFAQDYPEGT 443
R LIQL PVLL+LL+NFLPSSEP+Y+LS+SYPYEH+ T RGV YYVK F + YP +
Sbjct: 270 RMLIQLLPVLLLLLLNFLPSSEPVYSLSRSYPYEHKFQTTRGVTYYVKLPNFEEQYPHQS 329
Query: 442 RERETVEDKVEREYFGILRQNCQFELQRRQCGL 344
ER T+E VER+YF IL QNC+ E+QRR GL
Sbjct: 330 TERATLERHVERDYFSILSQNCRVEVQRRHWGL 362
Score = 29.3 bits (64), Expect(2) = 2e-30
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = -3
Query: 348 GYIKETPHCDILRKYE 301
G ETPHCD+LRK+E
Sbjct: 361 GLSYETPHCDMLRKFE 376
>ref|NP_199717.1| DNAJ-like protein; protein id: At5g49060.1 [Arabidopsis thaliana]
gi|10176939|dbj|BAB10088.1| DNAJ-like protein
[Arabidopsis thaliana]
Length = 350
Score = 61.2 bits (147), Expect = 1e-08
Identities = 33/90 (36%), Positives = 56/90 (61%), Gaps = 2/90 (2%)
Frame = -2
Query: 616 LIQLFPVLLILLINFLPSSEPIYTLSQSYPYEHRVTTPR-GVNYYVKS-TKFAQDYPEGT 443
+IQ+ P L+LL+ +LP SEP Y+L ++ Y+ TT +++YV+S + F + +P +
Sbjct: 240 IIQILPFFLLLLLAYLPFSEPDYSLHKNQSYQIPKTTQNTEISFYVRSASAFDEKFPLSS 299
Query: 442 RERETVEDKVEREYFGILRQNCQFELQRRQ 353
R +E V +EY L Q+C+ ELQ+R+
Sbjct: 300 SARANLEGNVIKEYKHFLFQSCRIELQKRR 329
>ref|NP_060096.1| DnaJ (Hsp40) homolog, subfamily B, member 12 [Homo sapiens]
gi|18203324|sp|Q9NXW2|DJBC_HUMAN DnaJ homolog subfamily
B member 12 gi|7019854|dbj|BAA90896.1| unnamed protein
product [Homo sapiens]
Length = 375
Score = 52.8 bits (125), Expect = 4e-06
Identities = 37/98 (37%), Positives = 55/98 (55%), Gaps = 6/98 (6%)
Frame = -2
Query: 613 IQLFPVLLILLINFLPS---SEPIYTLSQSYPYEH---RVTTPRGVNYYVKSTKFAQDYP 452
+QL P+L+++L++ L S P Y+LS H RVT GV YYV T F+++Y
Sbjct: 248 VQLMPILILILVSALSQLMVSSPPYSLSPRPSVGHIHRRVTDHLGVVYYVGDT-FSEEYT 306
Query: 451 EGTRERETVEDKVEREYFGILRQNCQFELQRRQCGLYQ 338
+ + TVE VE +Y LR NC E Q+++ LY+
Sbjct: 307 GSSLK--TVERNVEDDYIANLRNNCWKEKQQKEGLLYR 342
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 490,591,367
Number of Sequences: 1393205
Number of extensions: 9893389
Number of successful extensions: 26278
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 25217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26253
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)