Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001811A_C01 KMC001811A_c01
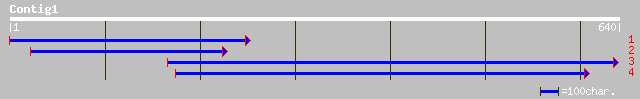
(637 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB90173.1| putative ATP-dependent Clp protease regulatory s... 134 1e-30
ref|NP_564423.1| ATP-dependent Clp protease ATP-binding subunit ... 126 3e-28
pir||D86457 hypothetical protein F10C21.5 - Arabidopsis thaliana... 126 3e-28
ref|NP_568714.1| ATP-dependent Clp protease ATP-binding subunit ... 118 7e-26
dbj|BAA98151.1| CLP protease regulatory subunit CLPX-like [Arabi... 118 7e-26
>dbj|BAB90173.1| putative ATP-dependent Clp protease regulatory subunit CLPX [Oryza
sativa (japonica cultivar-group)]
gi|21328048|dbj|BAC00633.1| putative ATP-dependent Clp
protease regulatory subunit CLPX [Oryza sativa (japonica
cultivar-group)]
Length = 625
Score = 134 bits (336), Expect = 1e-30
Identities = 65/113 (57%), Positives = 88/113 (77%), Gaps = 2/113 (1%)
Frame = -1
Query: 637 LFSMNNVKXHFTQKALRLIAKKAMAKNTGARGLRALMESILTEAMFEIPDVQAENELIDA 458
+FS+N VK HFT ALR++AKKA+A+NTGARGLRA++ES+L EAM+EIPD + +E +DA
Sbjct: 504 MFSLNKVKLHFTDGALRIVAKKAIARNTGARGLRAILESLLLEAMYEIPDEKTGSERVDA 563
Query: 457 VVVDEESVGSVDAPGCGGKILLGGGALEQYL--AKMEESVVNGGGAEPDLQES 305
VVVDEE++GS+D PGCG KIL G GALEQY+ M+ S+ G +L+++
Sbjct: 564 VVVDEEAIGSIDRPGCGAKILRGDGALEQYITNTNMKNSMEANEGLAGELEDA 616
>ref|NP_564423.1| ATP-dependent Clp protease ATP-binding subunit ClpX, putative;
protein id: At1g33360.1 [Arabidopsis thaliana]
gi|12322572|gb|AAG51286.1|AC027035_9 CLP protease
regulatory subunit CLPX, putative [Arabidopsis thaliana]
Length = 650
Score = 126 bits (316), Expect = 3e-28
Identities = 64/93 (68%), Positives = 76/93 (80%)
Frame = -1
Query: 637 LFSMNNVKXHFTQKALRLIAKKAMAKNTGARGLRALMESILTEAMFEIPDVQAENELIDA 458
LFSMNNVK HFT+KAL +I+K+AM KNTGARGLRAL+ESILTEAMFEIPD + +E IDA
Sbjct: 547 LFSMNNVKLHFTEKALEIISKQAMVKNTGARGLRALLESILTEAMFEIPDDKKGDERIDA 606
Query: 457 VVVDEESVGSVDAPGCGGKILLGGGALEQYLAK 359
V+VDEES S + GC KIL G GA E+YL++
Sbjct: 607 VIVDEESTSSEASRGCTAKILRGDGAFERYLSE 639
>pir||D86457 hypothetical protein F10C21.5 - Arabidopsis thaliana
gi|12322385|gb|AAG51217.1|AC051630_14 CLP protease
regulatory subunit CLPX, putative; 15869-19379
[Arabidopsis thaliana]
Length = 670
Score = 126 bits (316), Expect = 3e-28
Identities = 64/93 (68%), Positives = 76/93 (80%)
Frame = -1
Query: 637 LFSMNNVKXHFTQKALRLIAKKAMAKNTGARGLRALMESILTEAMFEIPDVQAENELIDA 458
LFSMNNVK HFT+KAL +I+K+AM KNTGARGLRAL+ESILTEAMFEIPD + +E IDA
Sbjct: 567 LFSMNNVKLHFTEKALEIISKQAMVKNTGARGLRALLESILTEAMFEIPDDKKGDERIDA 626
Query: 457 VVVDEESVGSVDAPGCGGKILLGGGALEQYLAK 359
V+VDEES S + GC KIL G GA E+YL++
Sbjct: 627 VIVDEESTSSEASRGCTAKILRGDGAFERYLSE 659
>ref|NP_568714.1| ATP-dependent Clp protease ATP-binding subunit ClpX, putative;
protein id: At5g49840.1 [Arabidopsis thaliana]
Length = 606
Score = 118 bits (295), Expect = 7e-26
Identities = 60/96 (62%), Positives = 77/96 (79%), Gaps = 1/96 (1%)
Frame = -1
Query: 637 LFSMNNVKXHFTQKALRLIAKKAMAKNTGARGLRALMESILTEAMFEIPDVQAE-NELID 461
LF MNNV+ FT+ A RLIA+KAM+KNTGARGLR+++ESILTEAMFE+PD E ++ I
Sbjct: 495 LFRMNNVQLQFTEGATRLIARKAMSKNTGARGLRSILESILTEAMFEVPDSITEGSQSIK 554
Query: 460 AVVVDEESVGSVDAPGCGGKILLGGGALEQYLAKME 353
AV+VDEE+VGSV +PGCG KIL G L+Q++ + E
Sbjct: 555 AVLVDEEAVGSVGSPGCGAKILKGDNVLQQFVEEAE 590
>dbj|BAA98151.1| CLP protease regulatory subunit CLPX-like [Arabidopsis thaliana]
Length = 608
Score = 118 bits (295), Expect = 7e-26
Identities = 60/96 (62%), Positives = 77/96 (79%), Gaps = 1/96 (1%)
Frame = -1
Query: 637 LFSMNNVKXHFTQKALRLIAKKAMAKNTGARGLRALMESILTEAMFEIPDVQAE-NELID 461
LF MNNV+ FT+ A RLIA+KAM+KNTGARGLR+++ESILTEAMFE+PD E ++ I
Sbjct: 497 LFRMNNVQLQFTEGATRLIARKAMSKNTGARGLRSILESILTEAMFEVPDSITEGSQSIK 556
Query: 460 AVVVDEESVGSVDAPGCGGKILLGGGALEQYLAKME 353
AV+VDEE+VGSV +PGCG KIL G L+Q++ + E
Sbjct: 557 AVLVDEEAVGSVGSPGCGAKILKGDNVLQQFVEEAE 592
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,014,313
Number of Sequences: 1393205
Number of extensions: 11538086
Number of successful extensions: 39634
Number of sequences better than 10.0: 284
Number of HSP's better than 10.0 without gapping: 35124
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39083
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)