Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001809A_C01 KMC001809A_c01
(538 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
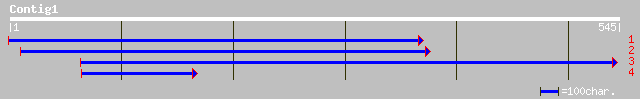
ref|NP_565804.1| expressed protein; protein id: At2g35470.1, sup... 63 3e-09
gb|AAL67580.1|AC018929_2 hypothetical protein [Oryza sativa] 47 1e-04
gb|AAL39351.1| GH26090p [Drosophila melanogaster] 35 0.57
ref|NP_611737.2| CG30190-PA [Drosophila melanogaster] gi|2162657... 35 0.57
emb|CAB37867.1| atrophin-1 [Hylobates lar] 34 1.3
>ref|NP_565804.1| expressed protein; protein id: At2g35470.1, supported by cDNA:
13821. [Arabidopsis thaliana] gi|25370787|pir||A84769
hypothetical protein At2g35470 [imported] - Arabidopsis
thaliana gi|3608141|gb|AAC36174.1| expressed protein
[Arabidopsis thaliana]
Length = 167
Score = 62.8 bits (151), Expect = 3e-09
Identities = 49/123 (39%), Positives = 63/123 (50%), Gaps = 23/123 (18%)
Frame = -1
Query: 505 KPHRLSSPLPAP-------PHKFIHIIPVLAVLCFLILFLCSHTPSPSDLDNFAGFKHSR 347
KP RLSS L +P K +H IP+L ++CF+IL+L S+ PS SDL F GF
Sbjct: 54 KPRRLSS-LQSPFVTTNQKQEKLVHFIPILTLICFIILYLTSYAPSQSDLAQFNGFMRP- 111
Query: 346 NPHLTDS---ATEISGGIEHYMEAKRSDVLAIR------------TLQQIPKPRL-HRKL 215
+ HL S EISG I R+D L+IR T + +P+ R HRK
Sbjct: 112 SKHLESSDENGDEISGFI-------RADTLSIRSSVRNLQETESFTTKSLPRRRTSHRKT 164
Query: 214 ADF 206
ADF
Sbjct: 165 ADF 167
>gb|AAL67580.1|AC018929_2 hypothetical protein [Oryza sativa]
Length = 155
Score = 47.0 bits (110), Expect = 1e-04
Identities = 21/33 (63%), Positives = 25/33 (75%)
Frame = -1
Query: 475 APPHKFIHIIPVLAVLCFLILFLCSHTPSPSDL 377
A P + IHIIPVL +LCFL+LFL SH P+ S L
Sbjct: 38 AAPDRSIHIIPVLTLLCFLVLFLLSHDPASSSL 70
>gb|AAL39351.1| GH26090p [Drosophila melanogaster]
Length = 292
Score = 35.0 bits (79), Expect = 0.57
Identities = 20/51 (39%), Positives = 26/51 (50%)
Frame = -3
Query: 530 RHHRRRTSQASPSVLASSRTATQVHPHHSSSRCPLFPHPLPLLSHSLSIRP 378
+H R +A PSV AS + + +H R P P PLP HSLS +P
Sbjct: 192 KHSFRAQLEAEPSVSASPSPSLRPKANHGKGRAP--PPPLPPKKHSLSPQP 240
>ref|NP_611737.2| CG30190-PA [Drosophila melanogaster] gi|21626579|gb|AAF46929.2|
CG30190-PA [Drosophila melanogaster]
Length = 408
Score = 35.0 bits (79), Expect = 0.57
Identities = 20/51 (39%), Positives = 26/51 (50%)
Frame = -3
Query: 530 RHHRRRTSQASPSVLASSRTATQVHPHHSSSRCPLFPHPLPLLSHSLSIRP 378
+H R +A PSV AS + + +H R P P PLP HSLS +P
Sbjct: 308 KHSFRAQLEAEPSVSASPSPSLRPKANHGKGRAP--PPPLPPKKHSLSPQP 356
>emb|CAB37867.1| atrophin-1 [Hylobates lar]
Length = 301
Score = 33.9 bits (76), Expect = 1.3
Identities = 23/52 (44%), Positives = 28/52 (53%)
Frame = -2
Query: 537 TPPSSPPTDLPSLTVCPRLFPHRHTSSSTSFQFSLSSVSSSSSSALTLPLHQ 382
+P S PP S P FP+ +SSS+S + SS SSSSSSA P Q
Sbjct: 16 SPHSLPPASSSSAPAPPMRFPY--SSSSSSSAAASSSSSSSSSSASPYPASQ 65
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 494,341,183
Number of Sequences: 1393205
Number of extensions: 11398261
Number of successful extensions: 61748
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 45401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 58499
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18173652336
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)