Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001807A_C01 KMC001807A_c01
(716 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
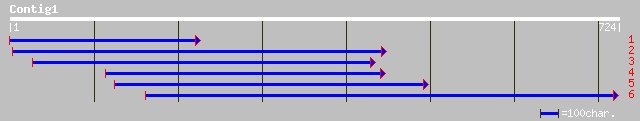
gb|AAL73489.1|AF464906_1 repressor protein [Glycine max] 236 2e-61
pir||S53582 TATA-binding protein-associated phosphoprotein Dr1 -... 194 8e-49
ref|NP_197700.1| TATA-binding protein-associated phosphoprotein ... 194 8e-49
pir||T50504 DR1-like protein - Arabidopsis thaliana gi|8346556|e... 192 4e-48
ref|NP_568190.1| DR1-like protein; protein id: At5g08190.1, supp... 187 1e-46
>gb|AAL73489.1|AF464906_1 repressor protein [Glycine max]
Length = 156
Score = 236 bits (602), Expect = 2e-61
Identities = 120/129 (93%), Positives = 124/129 (96%), Gaps = 1/129 (0%)
Frame = -1
Query: 716 MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVCGREERRTIAPEHVLKALGVLGFGDY 537
MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVC +EERRTIAPEHVLKALGVLGFG+Y
Sbjct: 28 MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVCNKEERRTIAPEHVLKALGVLGFGEY 87
Query: 536 IEEVYAAYEQHKLETM-DTLKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGGTITSKE 360
IEEVYAAYEQHKLETM D+LKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGG I SKE
Sbjct: 88 IEEVYAAYEQHKLETMQDSLKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGGAIQSKE 147
Query: 359 PDADQS*ES 333
P+ADQS ES
Sbjct: 148 PEADQSLES 156
>pir||S53582 TATA-binding protein-associated phosphoprotein Dr1 - Arabidopsis
thaliana
Length = 159
Score = 194 bits (494), Expect = 8e-49
Identities = 99/127 (77%), Positives = 109/127 (84%), Gaps = 3/127 (2%)
Frame = -1
Query: 716 MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVCGREERRTIAPEHVLKALGVLGFGDY 537
MLPPDVRVARDAQDLLIECCVEFINLVSSESN+VC +E++RTIAPEHVLKAL VLGFG+Y
Sbjct: 28 MLPPDVRVARDAQDLLIECCVEFINLVSSESNDVCNKEDKRTIAPEHVLKALQVLGFGEY 87
Query: 536 IEEVYAAYEQHKLETM-DTLKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGGTIT--S 366
IEEVYAAYEQHK ETM DT + KW+ A+MTEEEA AEQQRMFAEARARMNGG
Sbjct: 88 IEEVYAAYEQHKYETMQDTQRSVKWNPGAQMTEEEAAAEQQRMFAEARARMNGGVSVPQP 147
Query: 365 KEPDADQ 345
+ P+ DQ
Sbjct: 148 EHPETDQ 154
>ref|NP_197700.1| TATA-binding protein-associated phosphoprotein Dr1 protein homolog
(sp|P49592); protein id: At5g23090.1, supported by cDNA:
gi_16323209 [Arabidopsis thaliana]
gi|1352316|sp|P49592|DR1_ARATH Dr1 protein homolog
gi|633026|dbj|BAA07288.1| Dr1 [Arabidopsis thaliana]
gi|9759367|dbj|BAB09826.1| TATA-binding
protein-associated phosphoprotein Dr1 protein homolog
[Arabidopsis thaliana] gi|16323210|gb|AAL15339.1|
AT5g23090/MYJ24_8 [Arabidopsis thaliana]
gi|21436033|gb|AAM51594.1| AT5g23090/MYJ24_8
[Arabidopsis thaliana]
Length = 159
Score = 194 bits (494), Expect = 8e-49
Identities = 99/127 (77%), Positives = 109/127 (84%), Gaps = 3/127 (2%)
Frame = -1
Query: 716 MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVCGREERRTIAPEHVLKALGVLGFGDY 537
MLPPDVRVARDAQDLLIECCVEFINLVSSESN+VC +E++RTIAPEHVLKAL VLGFG+Y
Sbjct: 28 MLPPDVRVARDAQDLLIECCVEFINLVSSESNDVCNKEDKRTIAPEHVLKALQVLGFGEY 87
Query: 536 IEEVYAAYEQHKLETM-DTLKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGGTIT--S 366
IEEVYAAYEQHK ETM DT + KW+ A+MTEEEA AEQQRMFAEARARMNGG
Sbjct: 88 IEEVYAAYEQHKYETMQDTQRSVKWNPGAQMTEEEAAAEQQRMFAEARARMNGGVSVPQP 147
Query: 365 KEPDADQ 345
+ P+ DQ
Sbjct: 148 EHPETDQ 154
>pir||T50504 DR1-like protein - Arabidopsis thaliana gi|8346556|emb|CAB93720.1|
DR1-like protein [Arabidopsis thaliana]
Length = 162
Score = 192 bits (488), Expect = 4e-48
Identities = 95/121 (78%), Positives = 108/121 (88%)
Frame = -1
Query: 716 MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVCGREERRTIAPEHVLKALGVLGFGDY 537
MLP DVRVARDAQDLLIECCVEFINL+SSESNEVC +E++RTIAPEHVLKAL VLGFG+Y
Sbjct: 28 MLPADVRVARDAQDLLIECCVEFINLISSESNEVCNKEDKRTIAPEHVLKALQVLGFGEY 87
Query: 536 IEEVYAAYEQHKLETMDTLKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGGTITSKEP 357
+EEVYAAYEQHK ETMD+ + K ++ AEMTEEEA AEQQRMFAEARARMNGG +T +P
Sbjct: 88 VEEVYAAYEQHKYETMDSQRSVKMNSGAEMTEEEAAAEQQRMFAEARARMNGG-VTVPQP 146
Query: 356 D 354
+
Sbjct: 147 E 147
>ref|NP_568190.1| DR1-like protein; protein id: At5g08190.1, supported by cDNA:
30536., supported by cDNA: gi_16226437 [Arabidopsis
thaliana] gi|16226438|gb|AAL16168.1|AF428400_1
AT5g08190/T22D6_130 [Arabidopsis thaliana]
gi|21592629|gb|AAM64578.1| DR1-like protein [Arabidopsis
thaliana] gi|21928051|gb|AAM78054.1| AT5g08190/T22D6_130
[Arabidopsis thaliana]
Length = 163
Score = 187 bits (476), Expect = 1e-46
Identities = 95/122 (77%), Positives = 108/122 (87%), Gaps = 1/122 (0%)
Frame = -1
Query: 716 MLPPDVRVARDAQDLLIECCVEFINLVSSESNEVCGREERRTIAPEHVLKALGVLGFGDY 537
MLP DVRVARDAQDLLIECCVEFINL+SSESNEVC +E++RTIAPEHVLKAL VLGFG+Y
Sbjct: 28 MLPADVRVARDAQDLLIECCVEFINLISSESNEVCNKEDKRTIAPEHVLKALQVLGFGEY 87
Query: 536 IEEVYAAYEQHKLETM-DTLKGAKWSNRAEMTEEEALAEQQRMFAEARARMNGGTITSKE 360
+EEVYAAYEQHK ETM D+ + K ++ AEMTEEEA AEQQRMFAEARARMNGG +T +
Sbjct: 88 VEEVYAAYEQHKYETMQDSQRSVKMNSGAEMTEEEAAAEQQRMFAEARARMNGG-VTVPQ 146
Query: 359 PD 354
P+
Sbjct: 147 PE 148
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 567,926,573
Number of Sequences: 1393205
Number of extensions: 11405544
Number of successful extensions: 25690
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 24855
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25659
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33217548346
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)