Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
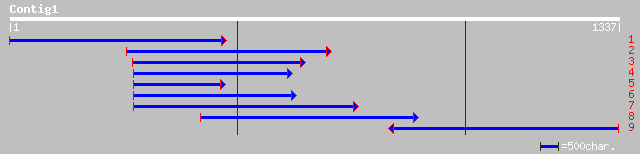
Query= KMC001775A_C01 KMC001775A_c01
(1296 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181435.1| unknown protein; protein id: At2g39020.1 [Arabi... 267 2e-70
ref|NP_565898.1| expressed protein; protein id: At2g39030.1, sup... 259 5e-68
gb|AAK96852.1| Unknown protein [Arabidopsis thaliana] gi|1837743... 254 2e-66
sp|Q9SMB8|THTB_TOBAC Tyramine N-feruloyltransferase 4/11 (Hydrox... 153 7e-36
emb|CAB55501.1| tyramine hydroxycinnamoyltransferase [Nicotiana ... 153 7e-36
>ref|NP_181435.1| unknown protein; protein id: At2g39020.1 [Arabidopsis thaliana]
gi|25408632|pir||B84812 hypothetical protein At2g39020
[imported] - Arabidopsis thaliana
gi|3928086|gb|AAC79612.1| unknown protein [Arabidopsis
thaliana] gi|28466877|gb|AAO44047.1| At2g39020
[Arabidopsis thaliana]
Length = 236
Score = 267 bits (683), Expect = 2e-70
Identities = 134/204 (65%), Positives = 157/204 (76%)
Frame = -2
Query: 1226 PMFTRIRLATPSDVPHILKMT*QLAVFEHFTHLFVGTESSLSSSLFSPNVQPFHSSTVFI 1047
PMF+RIRLATPSDVP I K+ Q+AVFE THLF TES L+S+LF+ +PF S TVF+
Sbjct: 29 PMFSRIRLATPSDVPFIHKLIHQMAVFERLTHLFSATESGLASTLFTS--RPFQSFTVFL 86
Query: 1046 LEASQNPFTDDTHLDNDPFYKPTMKVVNLDLIIDDPEKETFRNQHGNDIFMVGFVLFFPN 867
LE S++PF P + P K NLDL IDDPE F ND+ + GFVLFFPN
Sbjct: 87 LEVSRSPFPATITSSPSPDFTPFFKTHNLDLPIDDPESYNFSPDMLNDVVVAGFVLFFPN 146
Query: 866 YSTFMAKAGFYVEDLFVRECYRRKGFGKMLLSAVAKQAVKMGYGRVEWIVLDWNVNAIKF 687
YS+F++K GFY+ED+FVRE YRRKGFG MLL+AVAKQAVKMGYGRVEW+VLDWNVNAIKF
Sbjct: 147 YSSFLSKPGFYIEDIFVREPYRRKGFGSMLLTAVAKQAVKMGYGRVEWVVLDWNVNAIKF 206
Query: 686 YEGMGAKILQEWRLCRLIGDDLQA 615
YE MGA+ILQEWR+CRL GD L+A
Sbjct: 207 YEQMGAQILQEWRVCRLTGDALEA 230
>ref|NP_565898.1| expressed protein; protein id: At2g39030.1, supported by cDNA:
gi_15451161 [Arabidopsis thaliana]
gi|25408633|pir||C84812 hypothetical protein At2g39030
[imported] - Arabidopsis thaliana
gi|3928087|gb|AAC79613.1| expressed protein [Arabidopsis
thaliana]
Length = 228
Score = 259 bits (663), Expect = 5e-68
Identities = 132/226 (58%), Positives = 167/226 (73%)
Frame = -2
Query: 1292 PPRHIPNNSAAQRLPETARACQPMFTRIRLATPSDVPHILKMT*QLAVFEHFTHLFVGTE 1113
PP P + +PET+ MF+RIRLATP+DVP I K+ Q+AVFE THLFV TE
Sbjct: 3 PPTAAPEPNT---VPETSPTGHRMFSRIRLATPTDVPFIHKLIHQMAVFERLTHLFVATE 59
Query: 1112 SSLSSSLFSPNVQPFHSSTVFILEASQNPFTDDTHLDNDPFYKPTMKVVNLDLIIDDPEK 933
S L+S+LF N +PF + TVF+LE S +PF TH + P + P ++ +DL I+DP++
Sbjct: 60 SGLASTLF--NSRPFQAVTVFLLEISPSPFPT-THDASSPDFTPFLETHKVDLPIEDPDR 116
Query: 932 ETFRNQHGNDIFMVGFVLFFPNYSTFMAKAGFYVEDLFVRECYRRKGFGKMLLSAVAKQA 753
E F ND+ + GFVLFFPNY +F+AK GFY+ED+F+RE YRRKGFGK+LL+AVAKQA
Sbjct: 117 EKFLPDKLNDVVVAGFVLFFPNYPSFLAKQGFYIEDIFMREPYRRKGFGKLLLTAVAKQA 176
Query: 752 VKMGYGRVEWIVLDWNVNAIKFYEGMGAKILQEWRLCRLIGDDLQA 615
VK+G GRVEWIV+DWNVNAI FYE MGA++ +EWRLCRL GD LQA
Sbjct: 177 VKLGVGRVEWIVIDWNVNAINFYEQMGAQVFKEWRLCRLTGDALQA 222
>gb|AAK96852.1| Unknown protein [Arabidopsis thaliana] gi|18377432|gb|AAL66882.1|
unknown protein [Arabidopsis thaliana]
Length = 206
Score = 254 bits (649), Expect = 2e-66
Identities = 126/203 (62%), Positives = 158/203 (77%)
Frame = -2
Query: 1223 MFTRIRLATPSDVPHILKMT*QLAVFEHFTHLFVGTESSLSSSLFSPNVQPFHSSTVFIL 1044
MF+RIRLATP+DVP I K+ Q+AVFE THLFV TES L+S+LF N +PF + TVF+L
Sbjct: 1 MFSRIRLATPTDVPFIHKLIHQMAVFERLTHLFVATESGLASTLF--NSRPFQAVTVFLL 58
Query: 1043 EASQNPFTDDTHLDNDPFYKPTMKVVNLDLIIDDPEKETFRNQHGNDIFMVGFVLFFPNY 864
E S +PF TH + P + P ++ +DL I+DP++E F ND+ + GFVLFFPNY
Sbjct: 59 EISPSPFPT-THDASSPDFTPFLETHKVDLPIEDPDREKFLPDKLNDVVVAGFVLFFPNY 117
Query: 863 STFMAKAGFYVEDLFVRECYRRKGFGKMLLSAVAKQAVKMGYGRVEWIVLDWNVNAIKFY 684
+F+AK GFY+ED+F+RE YRRKGFGK+LL+AVAKQAVK+G GRVEWIV+DWNVNAI FY
Sbjct: 118 PSFLAKQGFYIEDIFMREPYRRKGFGKLLLTAVAKQAVKLGVGRVEWIVIDWNVNAINFY 177
Query: 683 EGMGAKILQEWRLCRLIGDDLQA 615
E MGA++ +EWRLCRL GD LQA
Sbjct: 178 EQMGAQVFKEWRLCRLTGDALQA 200
>sp|Q9SMB8|THTB_TOBAC Tyramine N-feruloyltransferase 4/11 (Hydroxycinnamoyl-CoA: tyramine
N-hydroxycinnamoyltransferase) gi|5852156|emb|CAB55502.1|
tyramine hydroxycinnamoyltransferase [Nicotiana tabacum]
Length = 226
Score = 153 bits (386), Expect = 7e-36
Identities = 80/215 (37%), Positives = 119/215 (55%), Gaps = 5/215 (2%)
Frame = -2
Query: 1229 QPMFTRIRLATPSDVPHILKMT*QLAVFEHFTHLFVGTESSLSSSLFSPNVQP-FHSSTV 1053
+ ++ R+RLA +D+ HI K+ Q+ + ++THL+ TESSL LF N P F+ +V
Sbjct: 13 EKVYVRVRLANEADISHIYKLFYQIHEYHNYTHLYKATESSLCDLLFKANPNPLFYGPSV 72
Query: 1052 FILEASQNPFTDDTHLDNDPFYKPTMKVVNLDLIIDDPEKETFRNQ----HGNDIFMVGF 885
+LE S PF + D +KP +K +L ++D E E F+++ D+F+ G+
Sbjct: 73 LLLEVSPTPFENTK---KDEKFKPVLKTFDLRATVEDKEAEEFKSKSCGDEKEDVFIAGY 129
Query: 884 VLFFPNYSTFMAKAGFYVEDLFVRECYRRKGFGKMLLSAVAKQAVKMGYGRVEWIVLDWN 705
F+ NYS F KAG Y E L+ RE YR+ G G +L VA A G+ VE IV WN
Sbjct: 130 AFFYANYSCFYDKAGIYFESLYFRESYRKLGMGSLLFGTVASIAANNGFASVEGIVAVWN 189
Query: 704 VNAIKFYEGMGAKILQEWRLCRLIGDDLQAHGGSD 600
+ FY MG +I E+R +L+GD LQ + +
Sbjct: 190 KKSYDFYVNMGVEIFDEFRYGKLVGDALQKYADKE 224
>emb|CAB55501.1| tyramine hydroxycinnamoyltransferase [Nicotiana tabacum]
Length = 233
Score = 153 bits (386), Expect = 7e-36
Identities = 80/215 (37%), Positives = 119/215 (55%), Gaps = 5/215 (2%)
Frame = -2
Query: 1229 QPMFTRIRLATPSDVPHILKMT*QLAVFEHFTHLFVGTESSLSSSLFSPNVQP-FHSSTV 1053
+ ++ R+RLA +D+ HI K+ Q+ + ++THL+ TESSL LF N P F+ +V
Sbjct: 20 EKVYVRVRLANEADISHIYKLFYQIHEYHNYTHLYKATESSLCDLLFKANPNPLFYGPSV 79
Query: 1052 FILEASQNPFTDDTHLDNDPFYKPTMKVVNLDLIIDDPEKETFRNQ----HGNDIFMVGF 885
+LE S PF + D +KP +K +L ++D E E F+++ D+F+ G+
Sbjct: 80 LLLEVSPTPFENTK---KDEKFKPVLKTFDLRATVEDKEAEEFKSKSCGDEKEDVFIAGY 136
Query: 884 VLFFPNYSTFMAKAGFYVEDLFVRECYRRKGFGKMLLSAVAKQAVKMGYGRVEWIVLDWN 705
F+ NYS F KAG Y E L+ RE YR+ G G +L VA A G+ VE IV WN
Sbjct: 137 AFFYANYSCFYDKAGIYFESLYFRESYRKLGMGSLLFGTVASIAANNGFASVEGIVAVWN 196
Query: 704 VNAIKFYEGMGAKILQEWRLCRLIGDDLQAHGGSD 600
+ FY MG +I E+R +L+GD LQ + +
Sbjct: 197 KKSYDFYVNMGVEIFDEFRYGKLVGDALQKYADKE 231
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,253,360,171
Number of Sequences: 1393205
Number of extensions: 32001500
Number of successful extensions: 136876
Number of sequences better than 10.0: 780
Number of HSP's better than 10.0 without gapping: 97548
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 127893
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 83309352185
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)