Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001769A_C01 KMC001769A_c01
(581 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
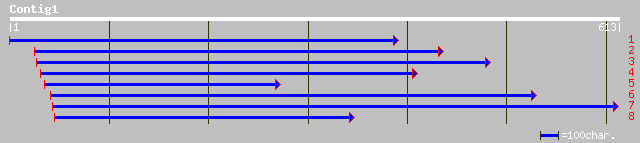
gb|AAL58201.1|AC090882_4 putative ATP(GTP)-binding protein [Oryz... 149 3e-35
ref|NP_567393.1| putative protein; protein id: At4g12790.1, supp... 139 3e-32
gb|EAA01032.1| agCP12481 [Anopheles gambiae str. PEST] 84 1e-15
ref|NP_649699.1| CG2656-PA [Drosophila melanogaster] gi|16768932... 80 2e-14
pir||T06637 hypothetical protein T20K18.140 - Arabidopsis thalia... 79 3e-14
>gb|AAL58201.1|AC090882_4 putative ATP(GTP)-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 279
Score = 149 bits (375), Expect = 3e-35
Identities = 70/95 (73%), Positives = 86/95 (89%)
Frame = -2
Query: 580 LSKMDLVTNKKDLDDFLDPEPTFLLSELNQRMAPQYAKLNKALIELVNNYSMVSFIPLDL 401
LSKMDLV+NKKD++++L+PE LLS+LN++MAP + KLNK+L ELV++YSMV+FIPLDL
Sbjct: 185 LSKMDLVSNKKDVEEYLNPEAQVLLSQLNRQMAPNFGKLNKSLAELVDDYSMVNFIPLDL 244
Query: 400 RKEKSIQYVLAQIDTCIQYGEDADVKVRDFDPEDD 296
RKE SIQYVL+ ID CIQYGEDADVKVRDFDPE+D
Sbjct: 245 RKESSIQYVLSHIDNCIQYGEDADVKVRDFDPEED 279
>ref|NP_567393.1| putative protein; protein id: At4g12790.1, supported by cDNA:
12532., supported by cDNA: gi_17065185 [Arabidopsis
thaliana] gi|17065186|gb|AAL32747.1| putative protein
[Arabidopsis thaliana] gi|21537254|gb|AAM61595.1|
putative ATP/GTP-binding protein [Arabidopsis thaliana]
gi|24899793|gb|AAN65111.1| putative protein [Arabidopsis
thaliana]
Length = 271
Score = 139 bits (349), Expect = 3e-32
Identities = 65/96 (67%), Positives = 82/96 (84%)
Frame = -2
Query: 580 LSKMDLVTNKKDLDDFLDPEPTFLLSELNQRMAPQYAKLNKALIELVNNYSMVSFIPLDL 401
LSKMDL+ +K ++DD+L+PEP LL+ELN+RM PQYAKLNKALIE+V Y MV+FIP++L
Sbjct: 172 LSKMDLLQDKSNIDDYLNPEPRTLLAELNKRMGPQYAKLNKALIEMVGEYGMVNFIPINL 231
Query: 400 RKEKSIQYVLAQIDTCIQYGEDADVKVRDFDPEDDE 293
RKEKSIQYVL+QID CIQ+GEDADV ++D D D+
Sbjct: 232 RKEKSIQYVLSQIDVCIQFGEDADVNIKDDDDFSDD 267
>gb|EAA01032.1| agCP12481 [Anopheles gambiae str. PEST]
Length = 322
Score = 84.0 bits (206), Expect = 1e-15
Identities = 44/100 (44%), Positives = 67/100 (67%), Gaps = 4/100 (4%)
Frame = -2
Query: 580 LSKMDLVT--NKKDLDDFLDPEPTFLLSELNQRMA--PQYAKLNKALIELVNNYSMVSFI 413
LSKMDL++ ++ +D +LDP+ LL E+ A +Y KL++ + L+ ++S+V F
Sbjct: 211 LSKMDLLSKVHRGQMDKYLDPDAHALLGEVTNESAWGRKYRKLSETIGMLIEDFSLVRFT 270
Query: 412 PLDLRKEKSIQYVLAQIDTCIQYGEDADVKVRDFDPEDDE 293
PL++ E+++ +L ID IQYGEDADVKVRDFDP + E
Sbjct: 271 PLNINDEENVADLLLMIDNVIQYGEDADVKVRDFDPPEPE 310
>ref|NP_649699.1| CG2656-PA [Drosophila melanogaster] gi|16768932|gb|AAL28685.1|
LD11854p [Drosophila melanogaster]
gi|23170590|gb|AAF54055.2| CG2656-PA [Drosophila
melanogaster]
Length = 283
Score = 79.7 bits (195), Expect = 2e-14
Identities = 43/101 (42%), Positives = 70/101 (68%), Gaps = 5/101 (4%)
Frame = -2
Query: 580 LSKMDLVTN--KKDLDDFLDPEPTFLLSELN--QRMAPQYAKLNKALIELVNNYSMVSFI 413
L+K+DL+++ +K L+ +L+P+ L+ EL +YAKL +A+ L+ ++S+V F
Sbjct: 181 LTKVDLLSSDARKQLEMYLEPDAHSLMGELTIGTGFGEKYAKLTQAIGALIEDFSLVRFF 240
Query: 412 PLDLRKEKSIQYVLAQIDTCIQYGEDADVKVRDFD-PEDDE 293
PLD + E+S+ +L QID+ +QYGEDADV V+DFD PE+ +
Sbjct: 241 PLDSQDEESVGDLLLQIDSILQYGEDADVNVKDFDEPEEGD 281
>pir||T06637 hypothetical protein T20K18.140 - Arabidopsis thaliana
gi|4586255|emb|CAB40996.1| putative protein [Arabidopsis
thaliana] gi|7267980|emb|CAB78321.1| putative protein
[Arabidopsis thaliana]
Length = 282
Score = 79.3 bits (194), Expect = 3e-14
Identities = 45/96 (46%), Positives = 60/96 (61%)
Frame = -2
Query: 580 LSKMDLVTNKKDLDDFLDPEPTFLLSELNQRMAPQYAKLNKALIELVNNYSMVSFIPLDL 401
LSKMDL+ +K ++D++ FL L +A V Y MV+FIP++L
Sbjct: 199 LSKMDLLQDKSNIDEY-----GFLFFPLFFSVAVS-----------VGEYGMVNFIPINL 242
Query: 400 RKEKSIQYVLAQIDTCIQYGEDADVKVRDFDPEDDE 293
RKEKSIQYVL+QID CIQ+GEDADV ++D D D+
Sbjct: 243 RKEKSIQYVLSQIDVCIQFGEDADVNIKDDDDFSDD 278
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 501,967,330
Number of Sequences: 1393205
Number of extensions: 10689176
Number of successful extensions: 24311
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 23540
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24299
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)