Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001765A_C01 KMC001765A_c01
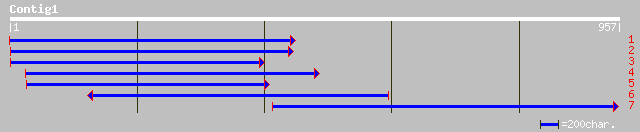
(957 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_174226.1| hypothetical protein; protein id: At1g29320.1 [... 234 1e-60
dbj|BAC16706.1| hypothetical protein~similar to Arabidopsis thal... 207 1e-52
ref|NP_598900.1| expressed sequence AA407588 [Mus musculus] gi|1... 90 4e-17
gb|AAH24478.1| Unknown (protein for MGC:37386) [Mus musculus] 90 4e-17
dbj|BAB15122.1| unnamed protein product [Homo sapiens] 89 1e-16
>ref|NP_174226.1| hypothetical protein; protein id: At1g29320.1 [Arabidopsis
thaliana]
Length = 468
Score = 234 bits (598), Expect = 1e-60
Identities = 131/236 (55%), Positives = 161/236 (67%), Gaps = 4/236 (1%)
Frame = -2
Query: 956 FLSKDDHRKFVAGTNSHQVRLYDISAQRRPVLSFDFRETPIKALAEDIDGYTVYIGNGSG 777
FLSKDDHRKFV GT SHQVRLYDIS QRRPVLSFDFRET I ++AED DG+T+Y+GN S
Sbjct: 222 FLSKDDHRKFVTGTKSHQVRLYDISTQRRPVLSFDFRETAITSIAEDPDGHTIYVGNASA 281
Query: 776 DMASVDIRTGKMLGGFTGKCSGSIRSIVRHPELPVIASCGLDSYLRIWDTKTRQLLSAVF 597
D+AS DIRTGK+LG F GKCSGSIRS+VRHP+ VIASCGLD YLR++D KTRQL+SAVF
Sbjct: 282 DLASFDIRTGKLLGSFLGKCSGSIRSVVRHPQHQVIASCGLDRYLRVYDVKTRQLISAVF 341
Query: 596 LKQPVLHALFDSSFIVEEPSSGADGLPSKAQNLNEIAAEE--EVEPSPLKRKKSSRNKEN 423
LKQ + +FDS F EE + + + I +E E E +P+KRKK
Sbjct: 342 LKQHLTGLVFDSGFSGEETAVANTVAEAATEEKMTIMDQEDDETEKAPVKRKK------- 394
Query: 422 NTNNSERKKRSKQSE-EGKKLKGNE-EHEKTASREKGSKSASKKRNKSSKRETSDE 261
S+++KRS++ EG+ + NE E EK + K SK + R K S+ E DE
Sbjct: 395 ----SKKEKRSREIVFEGEDDEENEDEIEKAPVKTKKSKKEKRSREKVSEGEEKDE 446
>dbj|BAC16706.1| hypothetical protein~similar to Arabidopsis thaliana chromosome1,
At1g29320 [Oryza sativa (japonica cultivar-group)]
Length = 462
Score = 207 bits (528), Expect = 1e-52
Identities = 118/238 (49%), Positives = 147/238 (61%), Gaps = 17/238 (7%)
Frame = -2
Query: 956 FLSKDDHRKFVAGTNSHQVRLYDISAQRRPVLSFDFRETPIKALAEDIDGYTVYIGNGSG 777
FL KDDHRK VA TN+HQVRLYD ++QRRPV+S DFRE+PIKA+AED +G+ VYIG G G
Sbjct: 236 FLCKDDHRKIVACTNNHQVRLYDTASQRRPVISVDFRESPIKAVAEDPNGHAVYIGTGRG 295
Query: 776 DMASVDIRT----------GKMLGGFTGKCSGSIRSIVRHPELPVIASC-------GLDS 648
D+AS D+RT GK+LG F GKCSGSIRSIVRHPELP+IASC GLDS
Sbjct: 296 DLASFDMRTELTLACTLFPGKLLGCFAGKCSGSIRSIVRHPELPLIASCGNQFTIVGLDS 355
Query: 647 YLRIWDTKTRQLLSAVFLKQPVLHALFDSSFIVEEPSSGADGLPSKAQNLNEIAAEEEVE 468
YLRIWDT TRQLLSAVFLKQ + + DS F EE +K++ + + AE
Sbjct: 356 YLRIWDTNTRQLLSAVFLKQHLTAVVIDSHFSTEELEE------TKSKQPDPVGAE---- 405
Query: 467 PSPLKRKKSSRNKENNTNNSERKKRSKQSEEGKKLKGNEEHEKTASREKGSKSASKKR 294
RK+ K + E + R ++ + +K+ + KG K SKK+
Sbjct: 406 ----VRKERKEKKNRTSEMDEDETRMLDHDDSDSEMHTSKRKKSGEKSKGMKKKSKKQ 459
>ref|NP_598900.1| expressed sequence AA407588 [Mus musculus]
gi|18043429|gb|AAH19968.1| Unknown (protein for
MGC:28494) [Mus musculus] gi|26346382|dbj|BAC36842.1|
unnamed protein product [Mus musculus]
Length = 384
Score = 90.1 bits (222), Expect = 4e-17
Identities = 57/155 (36%), Positives = 84/155 (53%), Gaps = 2/155 (1%)
Frame = -2
Query: 935 RKFVAGTNSHQVRLYD-ISAQRRPVLSFDFRETPIKALAEDIDGYTVYIGNGSGDMASVD 759
+K V T HQVR+YD +S QRRPVL + E P+ A+ +G +V +GN G +A +D
Sbjct: 196 QKLVTCTGYHQVRVYDPVSPQRRPVLEATYGEYPLTAMTLTPEGNSVIVGNTHGQLAEID 255
Query: 758 IRTGKMLGGFTGKCSGSIRSIVRHPELPVIASCGLDSYLRIWDTKT-RQLLSAVFLKQPV 582
R G++LG G +GS+R + HP P++ASCGLD LRI + R L V+LK +
Sbjct: 256 FRQGRLLGCLKG-LAGSVRGLQCHPSKPLLASCGLDRVLRIHRIRNPRGLEHKVYLKSQL 314
Query: 581 LHALFDSSFIVEEPSSGADGLPSKAQNLNEIAAEE 477
L E+ P + Q N++ +E+
Sbjct: 315 NCLLLSGRDNWEDE-------PQEPQEPNQVPSED 342
>gb|AAH24478.1| Unknown (protein for MGC:37386) [Mus musculus]
Length = 249
Score = 90.1 bits (222), Expect = 4e-17
Identities = 57/155 (36%), Positives = 84/155 (53%), Gaps = 2/155 (1%)
Frame = -2
Query: 935 RKFVAGTNSHQVRLYD-ISAQRRPVLSFDFRETPIKALAEDIDGYTVYIGNGSGDMASVD 759
+K V T HQVR+YD +S QRRPVL + E P+ A+ +G +V +GN G +A +D
Sbjct: 61 QKLVTCTGYHQVRVYDPVSPQRRPVLEATYGEYPLTAMTLTPEGNSVIVGNTHGQLAEID 120
Query: 758 IRTGKMLGGFTGKCSGSIRSIVRHPELPVIASCGLDSYLRIWDTKT-RQLLSAVFLKQPV 582
R G++LG G +GS+R + HP P++ASCGLD LRI + R L V+LK +
Sbjct: 121 FRQGRLLGCLKG-LAGSVRGLQCHPSKPLLASCGLDRVLRIHRIRNPRGLEHKVYLKSQL 179
Query: 581 LHALFDSSFIVEEPSSGADGLPSKAQNLNEIAAEE 477
L E+ P + Q N++ +E+
Sbjct: 180 NCLLLSGRDNWEDE-------PQEPQEPNQVPSED 207
>dbj|BAB15122.1| unnamed protein product [Homo sapiens]
Length = 250
Score = 88.6 bits (218), Expect = 1e-16
Identities = 63/192 (32%), Positives = 94/192 (48%), Gaps = 9/192 (4%)
Frame = -2
Query: 935 RKFVAGTNSHQVRLYD-ISAQRRPVLSFDFRETPIKALAEDIDGYTVYIGNGSGDMASVD 759
+K V T HQVR+YD S QRRPVL + E P+ A+ G +V +GN G +A +D
Sbjct: 61 QKLVTCTGYHQVRVYDPASPQRRPVLETTYGEYPLTAMTLTPGGNSVIVGNTHGQLAEID 120
Query: 758 IRTGKMLGGFTGKCSGSIRSIVRHPELPVIASCGLDSYLRIWDTKT-RQLLSAVFLKQPV 582
+R G++LG G +GS+R + HP P++ASCGLD LRI + R L V+LK +
Sbjct: 121 LRQGRLLGCLKG-LAGSVRGLQCHPSKPLLASCGLDRVLRIHRIQNPRGLEHKVYLKSQL 179
Query: 581 LHALFDSSFIVEEPSSGADGLPSKAQNLNEIAAEEEVE-------PSPLKRKKSSRNKEN 423
L E+ P + Q N++ E+ + KRK S +
Sbjct: 180 NCLLLSGRDNWEDE-------PQEPQEPNKVPLEDTETDELWASLEAAAKRKLSGLEQPQ 232
Query: 422 NTNNSERKKRSK 387
+ R+K+ +
Sbjct: 233 GALQTRRRKKKR 244
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 765,870,650
Number of Sequences: 1393205
Number of extensions: 16610886
Number of successful extensions: 112402
Number of sequences better than 10.0: 1923
Number of HSP's better than 10.0 without gapping: 70813
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 99204
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 54078381240
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)