Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001747A_C01 KMC001747A_c01
(704 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
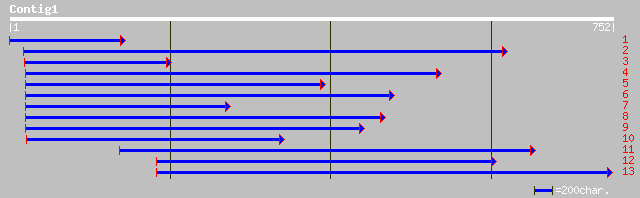
ref|NP_176678.1| unknown protein; protein id: At1g64980.1 [Arabi... 90 3e-17
gb|AAK39572.1|AC025296_7 unknown protein [Oryza sativa] 65 9e-10
dbj|BAC20854.1| hypothetical protein~similar to Oryza sativa chr... 62 6e-09
ref|NP_437086.1| conserved hypothetical protein [Sinorhizobium m... 35 1.3
gb|AAG41495.1| homeodomain protein DlxA [Petromyzon marinus] 33 3.8
>ref|NP_176678.1| unknown protein; protein id: At1g64980.1 [Arabidopsis thaliana]
gi|25373164|pir||D96673 hypothetical protein F13O11.28
[imported] - Arabidopsis thaliana
gi|5042435|gb|AAD38274.1|AC006193_30 Unknown protein
[Arabidopsis thaliana]
Length = 259
Score = 90.1 bits (222), Expect = 3e-17
Identities = 37/48 (77%), Positives = 42/48 (87%)
Frame = -2
Query: 703 VENDPTTLPKAIHYTRGGPWFEAWKHCEFADLWLNEMDEYLKQATKES 560
VE DPTT PKA+HYTRGGPWF+AWK CEFADLWLNEM+EY K+ KE+
Sbjct: 208 VEKDPTTQPKAVHYTRGGPWFDAWKDCEFADLWLNEMEEYNKENKKEA 255
>gb|AAK39572.1|AC025296_7 unknown protein [Oryza sativa]
Length = 268
Score = 65.1 bits (157), Expect = 9e-10
Identities = 25/44 (56%), Positives = 33/44 (74%)
Frame = -2
Query: 694 DPTTLPKAIHYTRGGPWFEAWKHCEFADLWLNEMDEYLKQATKE 563
DP+T PKAIHYT GGPWFE +++C+FA+LW+ E DE K+
Sbjct: 206 DPSTQPKAIHYTSGGPWFERYRNCDFAELWIKEADELKADKEKQ 249
>dbj|BAC20854.1| hypothetical protein~similar to Oryza sativa chromosome 10,
OSJNBa0076F20.4 [Oryza sativa (japonica cultivar-group)]
Length = 285
Score = 62.4 bits (150), Expect = 6e-09
Identities = 24/41 (58%), Positives = 32/41 (77%)
Frame = -2
Query: 685 TLPKAIHYTRGGPWFEAWKHCEFADLWLNEMDEYLKQATKE 563
T P+AIHYT GGPWFE +K+CEFA+LW+ E D Y +A ++
Sbjct: 220 TAPRAIHYTSGGPWFEQYKNCEFAELWVQERDAYEAEAEED 260
>ref|NP_437086.1| conserved hypothetical protein [Sinorhizobium meliloti]
gi|25373166|pir||B95910 conserved hypothetical protein
SMb20806 [imported] - Sinorhizobium meliloti (strain
1021) magaplasmid pSymB gi|15140431|emb|CAC48946.1|
conserved hypothetical protein [Sinorhizobium meliloti]
Length = 229
Score = 34.7 bits (78), Expect = 1.3
Identities = 12/32 (37%), Positives = 21/32 (65%)
Frame = -2
Query: 676 KAIHYTRGGPWFEAWKHCEFADLWLNEMDEYL 581
K +H+T GGP+F+ +K+ ++A+ W D L
Sbjct: 191 KLVHFTDGGPYFDEYKNDDYAEEWFATRDRVL 222
>gb|AAG41495.1| homeodomain protein DlxA [Petromyzon marinus]
Length = 400
Score = 33.1 bits (74), Expect = 3.8
Identities = 11/18 (61%), Positives = 15/18 (83%)
Frame = +3
Query: 186 YGLSYYHYHHNMHLHQMQ 239
YGLS++H+ H+ HLHQ Q
Sbjct: 24 YGLSHHHHQHHQHLHQQQ 41
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 561,458,381
Number of Sequences: 1393205
Number of extensions: 11225466
Number of successful extensions: 24552
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 22319
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24110
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32091529758
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)