Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001734A_C01 KMC001734A_c01
(505 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
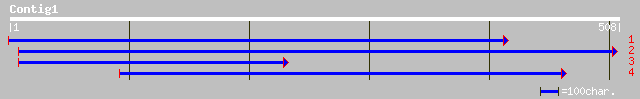
ref|NP_179402.1| putative elongation factor beta-1; protein id:... 164 4e-40
gb|AAM64977.1| putative elongation factor beta-1 [Arabidopsis t... 164 4e-40
sp|P48006|EF1B_ARATH Elongation factor 1-beta (EF-1-beta) 164 5e-40
ref|NP_174314.1| elongation factor 1-beta, putative; protein id:... 164 5e-40
sp|O81918|EF1B_BETVU ELONGATION FACTOR 1-BETA (EF-1-BETA) gi|744... 162 2e-39
>ref|NP_179402.1| putative elongation factor beta-1; protein id: At2g18110.1,
supported by cDNA: 35337., supported by cDNA:
gi_17473803, supported by cDNA: gi_20148478 [Arabidopsis
thaliana] gi|13124232|sp|Q9SI20|EF1C_ARATH Probable
elongation factor 1-beta (EF-1-beta)
gi|25299532|pir||D84560 probable elongation factor
1-beta [imported] - Arabidopsis thaliana
gi|4874292|gb|AAD31355.1| putative elongation factor
beta-1 [Arabidopsis thaliana] gi|17473804|gb|AAL38335.1|
putative elongation factor 1-beta [Arabidopsis thaliana]
gi|20148479|gb|AAM10130.1| putative elongation factor
1-beta [Arabidopsis thaliana] gi|20197598|gb|AAM15146.1|
putative elongation factor beta-1 [Arabidopsis
thaliana]
Length = 231
Score = 164 bits (416), Expect = 4e-40
Identities = 79/90 (87%), Positives = 85/90 (93%)
Frame = -1
Query: 505 KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGLFWGASKLVAVGYGIKKLQIMLTIVDDLV 326
KSSVL+D+KPWDDETDMKKLEEAVRS+Q EGLFWGASKLV VGYGIKKL IM TIVDDLV
Sbjct: 142 KSSVLMDIKPWDDETDMKKLEEAVRSIQMEGLFWGASKLVPVGYGIKKLHIMCTIVDDLV 201
Query: 325 SVDTLIEDHLTVEPINEYVQSCDIVAFNKI 236
S+DT+IE+ LTVEPINEYVQSCDIVAFNKI
Sbjct: 202 SIDTMIEEQLTVEPINEYVQSCDIVAFNKI 231
>gb|AAM64977.1| putative elongation factor beta-1 [Arabidopsis thaliana]
Length = 231
Score = 164 bits (416), Expect = 4e-40
Identities = 79/90 (87%), Positives = 85/90 (93%)
Frame = -1
Query: 505 KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGLFWGASKLVAVGYGIKKLQIMLTIVDDLV 326
KSSVL+D+KPWDDETDMKKLEEAVRS+Q EGLFWGASKLV VGYGIKKL IM TIVDDLV
Sbjct: 142 KSSVLMDIKPWDDETDMKKLEEAVRSIQMEGLFWGASKLVPVGYGIKKLHIMCTIVDDLV 201
Query: 325 SVDTLIEDHLTVEPINEYVQSCDIVAFNKI 236
S+DT+IE+ LTVEPINEYVQSCDIVAFNKI
Sbjct: 202 SIDTMIEEQLTVEPINEYVQSCDIVAFNKI 231
>sp|P48006|EF1B_ARATH Elongation factor 1-beta (EF-1-beta)
Length = 231
Score = 164 bits (415), Expect = 5e-40
Identities = 78/90 (86%), Positives = 86/90 (94%)
Frame = -1
Query: 505 KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGLFWGASKLVAVGYGIKKLQIMLTIVDDLV 326
KSSVL+D+KPWDDETDMKKLEEAV+S+Q EGLFWGASKLV VGYGIKKLQI+ TIVDDLV
Sbjct: 142 KSSVLIDIKPWDDETDMKKLEEAVKSIQMEGLFWGASKLVPVGYGIKKLQILCTIVDDLV 201
Query: 325 SVDTLIEDHLTVEPINEYVQSCDIVAFNKI 236
S+DT+IE+ LTVEPINEYVQSCDIVAFNKI
Sbjct: 202 SIDTMIEEQLTVEPINEYVQSCDIVAFNKI 231
>ref|NP_174314.1| elongation factor 1-beta, putative; protein id: At1g30230.1
[Arabidopsis thaliana] gi|25299531|pir||E86426 probable
elongation factor 1-beta [imported] - Arabidopsis
thaliana gi|12320854|gb|AAG50564.1|AC073506_6 elongation
factor 1-beta, putative [Arabidopsis thaliana]
Length = 242
Score = 164 bits (415), Expect = 5e-40
Identities = 78/90 (86%), Positives = 86/90 (94%)
Frame = -1
Query: 505 KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGLFWGASKLVAVGYGIKKLQIMLTIVDDLV 326
KSSVL+D+KPWDDETDMKKLEEAV+S+Q EGLFWGASKLV VGYGIKKLQI+ TIVDDLV
Sbjct: 142 KSSVLIDIKPWDDETDMKKLEEAVKSIQMEGLFWGASKLVPVGYGIKKLQILCTIVDDLV 201
Query: 325 SVDTLIEDHLTVEPINEYVQSCDIVAFNKI 236
S+DT+IE+ LTVEPINEYVQSCDIVAFNKI
Sbjct: 202 SIDTMIEEQLTVEPINEYVQSCDIVAFNKI 231
>sp|O81918|EF1B_BETVU ELONGATION FACTOR 1-BETA (EF-1-BETA) gi|7443457|pir||T14552
translation elongation factor eEF-1 beta chain homolog -
beet gi|3288113|emb|CAB09803.1| elongation factor 1-beta
[Beta vulgaris]
Length = 231
Score = 162 bits (410), Expect = 2e-39
Identities = 83/90 (92%), Positives = 85/90 (94%)
Frame = -1
Query: 505 KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGLFWGASKLVAVGYGIKKLQIMLTIVDDLV 326
KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGL GASKLV VGYGIKKL IM+TIVDDLV
Sbjct: 142 KSSVLLDVKPWDDETDMKKLEEAVRSVQQEGLTLGASKLVPVGYGIKKLTIMMTIVDDLV 201
Query: 325 SVDTLIEDHLTVEPINEYVQSCDIVAFNKI 236
SVD LIED+LTVEPINEYVQSCDIVAFNKI
Sbjct: 202 SVDNLIEDYLTVEPINEYVQSCDIVAFNKI 231
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 417,799,956
Number of Sequences: 1393205
Number of extensions: 8623822
Number of successful extensions: 25699
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 24003
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25435
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15362785481
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)