Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001726A_C01 KMC001726A_c01
(601 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
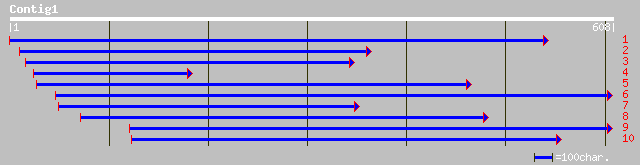
ref|NP_191593.1| palmitoyl-protein thioesterase precursor - like... 152 3e-36
gb|AAM65342.1| palmitoyl-protein thioesterase precursor-like [Ar... 152 3e-36
gb|AAK59537.1| putative palmitoyl-protein thioesterase precursor... 152 3e-36
ref|NP_193477.1| putative protein; protein id: At4g17470.1 [Arab... 132 3e-30
pir||C71444 probable thioesterase - Arabidopsis thaliana gi|2245... 128 6e-29
>ref|NP_191593.1| palmitoyl-protein thioesterase precursor - like; protein id:
At3g60340.1, supported by cDNA: 38539., supported by
cDNA: gi_14334717 [Arabidopsis thaliana]
gi|11358598|pir||T49229 palmitoyl-protein
thioesterase-like protein F27H5.130 [imported] -
Arabidopsis thaliana gi|7576210|emb|CAB87871.1|
palmitoyl-protein thioesterase precursor-like
[Arabidopsis thaliana] gi|23296336|gb|AAN13045.1|
putative palmitoyl-protein thioesterase precursor
[Arabidopsis thaliana]
Length = 338
Score = 152 bits (384), Expect = 3e-36
Identities = 71/93 (76%), Positives = 79/93 (84%)
Frame = -2
Query: 600 VLIMFEQDTVLIPKETAWFGYYPDGSFKPVLPPQETELYTEDWIGLRTLDEAGKVHFISV 421
VLIMFE DT+LIPKET+WFGYYPDGSFK +LPPQET+LYTEDWIGLRTLDEAGKV F++V
Sbjct: 210 VLIMFEHDTILIPKETSWFGYYPDGSFKTILPPQETKLYTEDWIGLRTLDEAGKVKFVNV 269
Query: 420 QGRHLGISEEDMQKHIVPYLKDQSSREYSTNKA 322
G HL IS DM+KHIVPYL D+SS ST A
Sbjct: 270 SGNHLQISHTDMKKHIVPYLCDKSSSSSSTTMA 302
>gb|AAM65342.1| palmitoyl-protein thioesterase precursor-like [Arabidopsis
thaliana]
Length = 338
Score = 152 bits (384), Expect = 3e-36
Identities = 71/93 (76%), Positives = 79/93 (84%)
Frame = -2
Query: 600 VLIMFEQDTVLIPKETAWFGYYPDGSFKPVLPPQETELYTEDWIGLRTLDEAGKVHFISV 421
VLIMFE DT+LIPKET+WFGYYPDGSFK +LPPQET+LYTEDWIGLRTLDEAGKV F++V
Sbjct: 210 VLIMFEHDTILIPKETSWFGYYPDGSFKTILPPQETKLYTEDWIGLRTLDEAGKVKFVNV 269
Query: 420 QGRHLGISEEDMQKHIVPYLKDQSSREYSTNKA 322
G HL IS DM+KHIVPYL D+SS ST A
Sbjct: 270 SGNHLQISHTDMKKHIVPYLCDKSSSSSSTTMA 302
>gb|AAK59537.1| putative palmitoyl-protein thioesterase precursor [Arabidopsis
thaliana]
Length = 338
Score = 152 bits (384), Expect = 3e-36
Identities = 71/93 (76%), Positives = 79/93 (84%)
Frame = -2
Query: 600 VLIMFEQDTVLIPKETAWFGYYPDGSFKPVLPPQETELYTEDWIGLRTLDEAGKVHFISV 421
VLIMFE DT+LIPKET+WFGYYPDGSFK +LPPQET+LYTEDWIGLRTLDEAGKV F++V
Sbjct: 210 VLIMFEHDTILIPKETSWFGYYPDGSFKTILPPQETKLYTEDWIGLRTLDEAGKVKFVNV 269
Query: 420 QGRHLGISEEDMQKHIVPYLKDQSSREYSTNKA 322
G HL IS DM+KHIVPYL D+SS ST A
Sbjct: 270 SGNHLQISHTDMKKHIVPYLCDKSSSSSSTTMA 302
>ref|NP_193477.1| putative protein; protein id: At4g17470.1 [Arabidopsis thaliana]
Length = 308
Score = 132 bits (333), Expect = 3e-30
Identities = 62/94 (65%), Positives = 80/94 (84%), Gaps = 4/94 (4%)
Frame = -2
Query: 600 VLIMFEQDTVLIPKETAWFGYYPDG--SFKPVLPPQETELYTEDWIGLRTLDEAGKVHFI 427
VLIMF+ DT+L+P+ET+WFGYYPDG S PVLPPQ+T+LYTEDWIGL+TLD+AGKV FI
Sbjct: 214 VLIMFQNDTILVPRETSWFGYYPDGASSSTPVLPPQKTKLYTEDWIGLKTLDDAGKVRFI 273
Query: 426 SVQGRHLGISEEDMQKHIVPYLKDQS--SREYST 331
SV G H+ I+EED+ K++VPYL+++S S E+ T
Sbjct: 274 SVPGGHIEITEEDLVKYVVPYLQNESLFSSEHET 307
>pir||C71444 probable thioesterase - Arabidopsis thaliana
gi|2245106|emb|CAB10528.1| thioesterase like protein
[Arabidopsis thaliana] gi|7268499|emb|CAB78750.1|
thioesterase like protein [Arabidopsis thaliana]
Length = 282
Score = 128 bits (321), Expect = 6e-29
Identities = 59/91 (64%), Positives = 77/91 (83%), Gaps = 4/91 (4%)
Frame = -2
Query: 591 MFEQDTVLIPKETAWFGYYPDG--SFKPVLPPQETELYTEDWIGLRTLDEAGKVHFISVQ 418
MF+ DT+L+P+ET+WFGYYPDG S PVLPPQ+T+LYTEDWIGL+TLD+AGKV FISV
Sbjct: 191 MFQNDTILVPRETSWFGYYPDGASSSTPVLPPQKTKLYTEDWIGLKTLDDAGKVRFISVP 250
Query: 417 GRHLGISEEDMQKHIVPYLKDQS--SREYST 331
G H+ I+EED+ K++VPYL+++S S E+ T
Sbjct: 251 GGHIEITEEDLVKYVVPYLQNESLFSSEHET 281
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,215,060
Number of Sequences: 1393205
Number of extensions: 9531194
Number of successful extensions: 21932
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 21486
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21920
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)