Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001725A_C01 KMC001725A_c01
(661 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
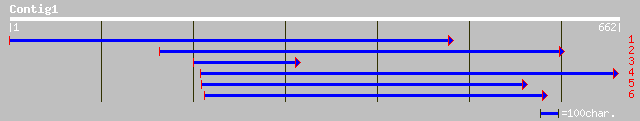
ref|NP_671924.1| similar to chloroplast nucleoid DNA binding pro... 72 9e-12
ref|NP_176663.1| hypothetical protein; protein id: At1g64830.1 [... 66 4e-10
ref|NP_174430.1| chloroplast nucleoid DNA binding protein, putat... 65 6e-10
dbj|BAB07973.1| P0406H10.5 [Oryza sativa (japonica cultivar-grou... 59 6e-08
ref|NP_198319.1| putative protein; protein id: At5g33340.1 [Arab... 57 3e-07
>ref|NP_671924.1| similar to chloroplast nucleoid DNA binding protein, putative;
protein id: At2g35615.1 [Arabidopsis thaliana]
Length = 447
Score = 71.6 bits (174), Expect = 9e-12
Identities = 34/83 (40%), Positives = 49/83 (58%)
Frame = -3
Query: 629 CFTYKEGMSTPPDVVFHFTGGDVVLKPMNTLVLIEDNLICSPVVPSHFDGIAIFGNLGQI 450
CF P++ HFTG DV L P+N V + ++++C +VP+ +AI+GN Q+
Sbjct: 366 CFKSGSAEIGLPEITVHFTGADVRLSPINAFVKLSEDMVCLSMVPT--TEVAIYGNFAQM 423
Query: 449 DFHVGYDIQGGKVSFAPTDCSLN 381
DF VGYD++ VSF DCS N
Sbjct: 424 DFLVGYDLETRTVSFQHMDCSAN 446
>ref|NP_176663.1| hypothetical protein; protein id: At1g64830.1 [Arabidopsis
thaliana] gi|25404498|pir||E96671 hypothetical protein
F13O11.13 [imported] - Arabidopsis thaliana
gi|5042418|gb|AAD38257.1|AC006193_13 Hypothetical
Protein [Arabidopsis thaliana]
Length = 431
Score = 66.2 bits (160), Expect = 4e-10
Identities = 32/70 (45%), Positives = 43/70 (60%)
Frame = -3
Query: 596 PDVVFHFTGGDVVLKPMNTLVLIEDNLICSPVVPSHFDGIAIFGNLGQIDFHVGYDIQGG 417
PD+ HF GGDV L +NT V + +++ C + + + IFGNL Q++F VGYD G
Sbjct: 362 PDITVHFKGGDVKLGNLNTFVAVSEDVSCFAFAAN--EQLTIFGNLAQMNFLVGYDTVSG 419
Query: 416 KVSFAPTDCS 387
VSF TDCS
Sbjct: 420 TVSFKKTDCS 429
>ref|NP_174430.1| chloroplast nucleoid DNA binding protein, putative; protein id:
At1g31450.1 [Arabidopsis thaliana]
gi|25513600|pir||E86440 probable chloroplast nucleoid
DNA binding protein T8E3.12 - Arabidopsis thaliana
gi|12322538|gb|AAG51267.1|AC027135_8 chloroplast
nucleoid DNA binding protein, putative [Arabidopsis
thaliana]
Length = 445
Score = 65.5 bits (158), Expect = 6e-10
Identities = 32/83 (38%), Positives = 48/83 (57%)
Frame = -3
Query: 629 CFTYKEGMSTPPDVVFHFTGGDVVLKPMNTLVLIEDNLICSPVVPSHFDGIAIFGNLGQI 450
CF + P + HFT DV L P+N V + ++ +C ++P+ +AI+GN+ Q+
Sbjct: 364 CFKSGDKEIGLPAITMHFTNADVKLSPINAFVKLNEDTVCLSMIPT--TEVAIYGNMVQM 421
Query: 449 DFHVGYDIQGGKVSFAPTDCSLN 381
DF VGYD++ VSF DCS N
Sbjct: 422 DFLVGYDLETKTVSFQRMDCSGN 444
>dbj|BAB07973.1| P0406H10.5 [Oryza sativa (japonica cultivar-group)]
gi|15289772|dbj|BAB63472.1| P0509B06.7 [Oryza sativa
(japonica cultivar-group)]
Length = 451
Score = 58.9 bits (141), Expect = 6e-08
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 2/72 (2%)
Frame = -3
Query: 596 PDVVFHFTGGDVV-LKPMNTLVLIEDNLICSPVVPS-HFDGIAIFGNLGQIDFHVGYDIQ 423
PD+ F GG V LKP N V +++ +C +V + ++I GNL Q + HVGYD+
Sbjct: 377 PDLTLEFGGGAAVALKPENAFVAVQEGTLCLAIVATTEQQPVSILGNLAQQNIHVGYDLD 436
Query: 422 GGKVSFAPTDCS 387
G V+FA DC+
Sbjct: 437 AGTVTFAGADCA 448
>ref|NP_198319.1| putative protein; protein id: At5g33340.1 [Arabidopsis thaliana]
Length = 437
Score = 56.6 bits (135), Expect = 3e-07
Identities = 28/70 (40%), Positives = 39/70 (55%)
Frame = -3
Query: 596 PDVVFHFTGGDVVLKPMNTLVLIEDNLICSPVVPSHFDGIAIFGNLGQIDFHVGYDIQGG 417
P + HF G DV L N V + ++L+C S +I+GN+ Q++F VGYD
Sbjct: 368 PVITMHFDGADVKLDSSNAFVQVSEDLVCFAFRGS--PSFSIYGNVAQMNFLVGYDTVSK 425
Query: 416 KVSFAPTDCS 387
VSF PTDC+
Sbjct: 426 TVSFKPTDCA 435
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 571,818,345
Number of Sequences: 1393205
Number of extensions: 12479028
Number of successful extensions: 29205
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 28244
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29177
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28289785200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)