Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
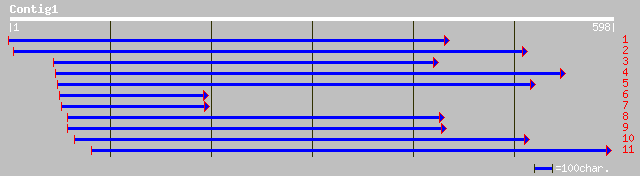
Query= KMC001719A_C01 KMC001719A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P16147|PERX_LUPPO Peroxidase gi|81844|pir||S26672 peroxidase ... 138 6e-32
gb|AAD43561.1|AF155124_1 bacterial-induced peroxidase precursor ... 130 1e-29
pir||T09164 probable peroxidase (EC 1.11.1.7) (clone PC44) - spi... 125 3e-28
pir||T09166 probable peroxidase (EC 1.11.1.7) (clone PC23) - spi... 124 9e-28
dbj|BAA77389.1| peroxidase 3 [Scutellaria baicalensis] 119 4e-26
>sp|P16147|PERX_LUPPO Peroxidase gi|81844|pir||S26672 peroxidase (EC 1.11.1.7) -
large-leaved lupine (fragment)
gi|1345541|emb|CAA36066.1| peroxidase [Lupinus
polyphyllus] gi|228535|prf||1805332A
peroxidase:ISOTYPE=basic isozyme
Length = 158
Score = 138 bits (347), Expect = 6e-32
Identities = 63/84 (75%), Positives = 75/84 (89%)
Frame = -3
Query: 584 APLETLTPTKFENNYYRDLVARKGLLHSDQVLFNGGSQDALVKSYAANNAAFFRDFAAAM 405
APL++LTP +F+NNYY+DLV+ +GLLHSDQVLFNGGSQD LV++Y+ NN FF DFAAA+
Sbjct: 75 APLDSLTPNRFDNNYYKDLVSNRGLLHSDQVLFNGGSQDTLVRTYSTNNVKFFSDFAAAI 134
Query: 404 VKMSKISPLTGTNGEIRKNCRVVN 333
VKMSKISPLTG GEIRKNCRV+N
Sbjct: 135 VKMSKISPLTGIAGEIRKNCRVIN 158
>gb|AAD43561.1|AF155124_1 bacterial-induced peroxidase precursor [Gossypium hirsutum]
Length = 316
Score = 130 bits (327), Expect = 1e-29
Identities = 62/84 (73%), Positives = 73/84 (86%)
Frame = -3
Query: 584 APLETLTPTKFENNYYRDLVARKGLLHSDQVLFNGGSQDALVKSYAANNAAFFRDFAAAM 405
APL+ TPT+F+N+Y+R+LVAR+GLLHSDQ LFNGGSQDALV++Y+ N A F DFAAAM
Sbjct: 233 APLDIQTPTRFDNDYFRNLVARRGLLHSDQELFNGGSQDALVRTYSNNPATFSADFAAAM 292
Query: 404 VKMSKISPLTGTNGEIRKNCRVVN 333
VKM ISPLTGT GEIR+NCRVVN
Sbjct: 293 VKMGNISPLTGTQGEIRRNCRVVN 316
>pir||T09164 probable peroxidase (EC 1.11.1.7) (clone PC44) - spinach
gi|1781328|emb|CAA71491.1| peroxidase [Spinacia
oleracea]
Length = 323
Score = 125 bits (315), Expect = 3e-28
Identities = 55/84 (65%), Positives = 71/84 (84%)
Frame = -3
Query: 584 APLETLTPTKFENNYYRDLVARKGLLHSDQVLFNGGSQDALVKSYAANNAAFFRDFAAAM 405
AP++ TP F+N+YY++LVA++GLLHSDQ L+NGGSQD+LVK Y+ N A FF+DFAAAM
Sbjct: 240 APMDIQTPNTFDNDYYKNLVAKRGLLHSDQELYNGGSQDSLVKMYSTNQALFFQDFAAAM 299
Query: 404 VKMSKISPLTGTNGEIRKNCRVVN 333
++M + PLTGTNGEIR NCRV+N
Sbjct: 300 IRMGDLKPLTGTNGEIRNNCRVIN 323
>pir||T09166 probable peroxidase (EC 1.11.1.7) (clone PC23) - spinach
(fragment) gi|1781332|emb|CAA71493.1| peroxidase
[Spinacia oleracea]
Length = 309
Score = 124 bits (311), Expect = 9e-28
Identities = 55/84 (65%), Positives = 73/84 (86%)
Frame = -3
Query: 584 APLETLTPTKFENNYYRDLVARKGLLHSDQVLFNGGSQDALVKSYAANNAAFFRDFAAAM 405
APL+ +PTKF+N+YY++L+A++GLLHSDQ L+NGGSQDALV Y+ +NAAF +DF AA+
Sbjct: 226 APLDLQSPTKFDNSYYKNLIAKRGLLHSDQELYNGGSQDALVTRYSKSNAAFAKDFVAAI 285
Query: 404 VKMSKISPLTGTNGEIRKNCRVVN 333
+KM ISPLTG++GEIRKNCR +N
Sbjct: 286 IKMGNISPLTGSSGEIRKNCRFIN 309
>dbj|BAA77389.1| peroxidase 3 [Scutellaria baicalensis]
Length = 318
Score = 119 bits (297), Expect = 4e-26
Identities = 53/84 (63%), Positives = 69/84 (82%)
Frame = -3
Query: 584 APLETLTPTKFENNYYRDLVARKGLLHSDQVLFNGGSQDALVKSYAANNAAFFRDFAAAM 405
APL+ ++P +F N+YYR+L+ +GLLHSDQ LFN G+ DA V++Y+ N+AAFF DFA AM
Sbjct: 235 APLDNVSPARFNNDYYRNLIGLRGLLHSDQELFNNGTADAQVRAYSTNSAAFFNDFANAM 294
Query: 404 VKMSKISPLTGTNGEIRKNCRVVN 333
VKMS +SPLTGTNG+IR+NCR N
Sbjct: 295 VKMSNLSPLTGTNGQIRRNCRRTN 318
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 468,522,109
Number of Sequences: 1393205
Number of extensions: 9370767
Number of successful extensions: 22352
Number of sequences better than 10.0: 525
Number of HSP's better than 10.0 without gapping: 21602
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22097
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)