Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001716A_C01 KMC001716A_c01
(935 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
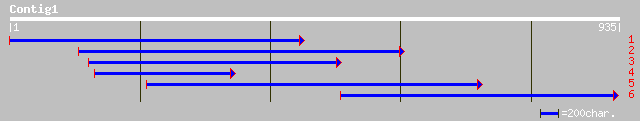
ref|NP_190390.1| putative protein; protein id: At3g48070.1, supp... 133 4e-44
ref|NP_201096.1| putative protein; protein id: At5g62910.1 [Arab... 131 1e-41
gb|AAL86287.1| unknown protein [Arabidopsis thaliana] 131 1e-41
emb|CAD41040.1| OSJNBa0060P14.11 [Oryza sativa (japonica cultiva... 95 1e-27
ref|NP_177625.1| hypothetical protein; protein id: At1g74870.1 [... 77 3e-13
>ref|NP_190390.1| putative protein; protein id: At3g48070.1, supported by cDNA:
122162. [Arabidopsis thaliana] gi|7487127|pir||T06676
hypothetical protein T17F15.60 - Arabidopsis thaliana
gi|4678321|emb|CAB41132.1| putative protein [Arabidopsis
thaliana] gi|21537154|gb|AAM61495.1| unknown
[Arabidopsis thaliana]
Length = 319
Score = 133 bits (335), Expect(2) = 4e-44
Identities = 59/79 (74%), Positives = 67/79 (84%)
Frame = -1
Query: 608 PSSCPICFEDLDLTDTSFLPCLCGFRLCLFCHKRILEDDGRCPGCRKPYEREPVETEASV 429
PSSCPIC+EDLDLTD++FLPC CGFRLCLFCHK I + DGRCPGCRKPYER ++ EASV
Sbjct: 241 PSSCPICYEDLDLTDSNFLPCPCGFRLCLFCHKTICDGDGRCPGCRKPYERNTIKFEASV 300
Query: 428 VGGSLTLRLARSCSMIERA 372
GG LT+RLARS SM R+
Sbjct: 301 QGGGLTIRLARSSSMFCRS 319
Score = 67.8 bits (164), Expect(2) = 4e-44
Identities = 44/111 (39%), Positives = 56/111 (49%), Gaps = 1/111 (0%)
Frame = -3
Query: 933 DDEG-LDDWEAVADALAADDKNQCPCSDLPPSHDEHVGQTVPPRAAANGSDSNSKPASGR 757
DD+G LDDWEA+ADALAADD+ + P S EH + + A G+D + A
Sbjct: 139 DDDGCLDDWEAIADALAADDEKH-EKENPPESCQEH--EDIKQSALIGGADVVLRDAKAD 195
Query: 756 LVPWGSGNVRAWRADDAFRPQSLPNLSKQHSMPNPDRYCGGVPWCSQFCTI 604
+ +AWR DD RPQ LPNL KQ S P + + V C I
Sbjct: 196 SQRRKQKSNQAWRPDDKLRPQGLPNLEKQRSFPVMNLHFSSVTVVPSSCPI 246
>ref|NP_201096.1| putative protein; protein id: At5g62910.1 [Arabidopsis thaliana]
gi|10177469|dbj|BAB10860.1|
gene_id:MQB2.23~pir||T06676~similar to unknown protein
[Arabidopsis thaliana]
Length = 327
Score = 131 bits (330), Expect(2) = 1e-41
Identities = 57/75 (76%), Positives = 64/75 (85%)
Frame = -1
Query: 608 PSSCPICFEDLDLTDTSFLPCLCGFRLCLFCHKRILEDDGRCPGCRKPYEREPVETEASV 429
PSSCPIC+EDLDLTD++FLPC CGFRLCLFCHK I + DGRCPGCRKPYER V+ E S+
Sbjct: 249 PSSCPICYEDLDLTDSNFLPCPCGFRLCLFCHKTICDGDGRCPGCRKPYERNMVKAETSI 308
Query: 428 VGGSLTLRLARSCSM 384
GG LT+RLARS SM
Sbjct: 309 QGGGLTIRLARSSSM 323
Score = 61.2 bits (147), Expect(2) = 1e-41
Identities = 44/110 (40%), Positives = 59/110 (53%), Gaps = 14/110 (12%)
Frame = -3
Query: 933 DDEG-LDDWEAVADALAADDKNQCPCSDLPPSHDE-HVGQTVPPRAAANGSDSNSKPASG 760
DD+G +DDWEAVADALAA+++ + L ++ VGQ+ A+N DS+ AS
Sbjct: 137 DDDGCVDDWEAVADALAAEEEIEKKSRPLESVKEQVSVGQS-----ASNVCDSSISDASD 191
Query: 759 ------------RLVPWGSGNVRAWRADDAFRPQSLPNLSKQHSMPNPDR 646
R+ + RAWR DD RPQ LPNL+KQ S P D+
Sbjct: 192 VVGVEDPKQECLRVSSRKQTSNRAWRLDDDLRPQGLPNLAKQLSFPELDK 241
>gb|AAL86287.1| unknown protein [Arabidopsis thaliana]
Length = 222
Score = 131 bits (330), Expect(2) = 1e-41
Identities = 57/75 (76%), Positives = 64/75 (85%)
Frame = -1
Query: 608 PSSCPICFEDLDLTDTSFLPCLCGFRLCLFCHKRILEDDGRCPGCRKPYEREPVETEASV 429
PSSCPIC+EDLDLTD++FLPC CGFRLCLFCHK I + DGRCPGCRKPYER V+ E S+
Sbjct: 144 PSSCPICYEDLDLTDSNFLPCPCGFRLCLFCHKTICDGDGRCPGCRKPYERNMVKAETSI 203
Query: 428 VGGSLTLRLARSCSM 384
GG LT+RLARS SM
Sbjct: 204 QGGGLTIRLARSSSM 218
Score = 61.2 bits (147), Expect(2) = 1e-41
Identities = 44/110 (40%), Positives = 59/110 (53%), Gaps = 14/110 (12%)
Frame = -3
Query: 933 DDEG-LDDWEAVADALAADDKNQCPCSDLPPSHDE-HVGQTVPPRAAANGSDSNSKPASG 760
DD+G +DDWEAVADALAA+++ + L ++ VGQ+ A+N DS+ AS
Sbjct: 32 DDDGCVDDWEAVADALAAEEEIEKKSRPLESVKEQVSVGQS-----ASNVCDSSISDASD 86
Query: 759 ------------RLVPWGSGNVRAWRADDAFRPQSLPNLSKQHSMPNPDR 646
R+ + RAWR DD RPQ LPNL+KQ S P D+
Sbjct: 87 VVGVEDPKQECLRVSSRKQTSNRAWRLDDDLRPQGLPNLAKQLSFPELDK 136
>emb|CAD41040.1| OSJNBa0060P14.11 [Oryza sativa (japonica cultivar-group)]
Length = 341
Score = 94.7 bits (234), Expect(2) = 1e-27
Identities = 45/77 (58%), Positives = 50/77 (64%)
Frame = -1
Query: 614 SAPSSCPICFEDLDLTDTSFLPCLCGFRLCLFCHKRILEDDGRCPGCRKPYEREPVETEA 435
S P +CPIC EDLDLTD+SF PC C F LCLFCH +ILE DGRCPGCRK Y
Sbjct: 276 STPLTCPICCEDLDLTDSSFCPCPCKFCLCLFCHNKILEADGRCPGCRKEY--------- 326
Query: 434 SVVGGSLTLRLARSCSM 384
+ RL+RSCSM
Sbjct: 327 ------VAARLSRSCSM 337
Score = 51.6 bits (122), Expect(2) = 1e-27
Identities = 37/90 (41%), Positives = 44/90 (48%), Gaps = 2/90 (2%)
Frame = -3
Query: 921 LDDWEAVADA--LAADDKNQCPCSDLPPSHDEHVGQTVPPRAAANGSDSNSKPASGRLVP 748
LDDWEAVADA L DD H V P AA N + + +GR P
Sbjct: 184 LDDWEAVADADALTVDD-----------CHSHQSSGHVAPPAAPNVCTAPANQ-TGRQDP 231
Query: 747 WGSGNVRAWRADDAFRPQSLPNLSKQHSMP 658
+AW DD FRPQSLP++S+Q S P
Sbjct: 232 --IQRTKAWAPDDIFRPQSLPSISRQVSFP 259
>ref|NP_177625.1| hypothetical protein; protein id: At1g74870.1 [Arabidopsis
thaliana] gi|25406390|pir||C96778 hypothetical protein
F9E10.28 [imported] - Arabidopsis thaliana
gi|5882736|gb|AAD55289.1|AC008263_20 F25A4.16
[Arabidopsis thaliana]
gi|12323896|gb|AAG51922.1|AC013258_16 hypothetical
protein; 76274-75092 [Arabidopsis thaliana]
Length = 289
Score = 77.4 bits (189), Expect = 3e-13
Identities = 37/80 (46%), Positives = 50/80 (62%), Gaps = 4/80 (5%)
Frame = -1
Query: 599 CPICFEDLDLTDTSFLPCLCGFRLCLFCHKRILEDDGRCPGCRKPYEREPVETEASVVG- 423
CPIC E +D TD F PC CGFR+CLFCH +I E++ RCP CRK Y++ ++ VG
Sbjct: 212 CPICSELMDATDLEFEPCTCGFRICLFCHNKISENEARCPACRKDYKK--TSKKSGEVGY 269
Query: 422 ---GSLTLRLARSCSMIERA 372
G T+ L+ S ++RA
Sbjct: 270 QQRGRGTIPLSPSFRGLDRA 289
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 873,210,385
Number of Sequences: 1393205
Number of extensions: 21316598
Number of successful extensions: 69318
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 63537
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 69025
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 52137106016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)