Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001703A_C01 KMC001703A_c01
(594 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
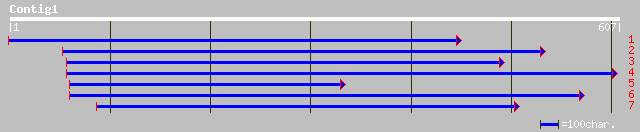
ref|NP_680171.1| similar to reverse transcriptase, putative; pro... 34 1.6
ref|NP_654146.1| DUF6, Integral membrane protein DUF6 [Bacillus ... 33 3.6
ref|NP_691788.1| hypothetical protein [Oceanobacillus iheyensis ... 33 3.6
ref|NP_207698.1| hypothetical protein [Helicobacter pylori 26695... 32 4.7
ref|NP_051962.1| gp073R [Rabbit fibroma virus] gi|6578601|gb|AAF... 32 6.1
>ref|NP_680171.1| similar to reverse transcriptase, putative; protein id: At5g17725.1
[Arabidopsis thaliana]
Length = 721
Score = 33.9 bits (76), Expect = 1.6
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 2/75 (2%)
Frame = +2
Query: 170 KVSELSRIH*EYTTNTWSKTSFTKVYPKLRGKK*KLETKSLIKEVTVIRRKGNSSKLFGE 349
+V +++I EY+ + K SF+K + G K + ET++LIKE+T I ++G + G
Sbjct: 59 EVDVINQILQEYSRVSGQKVSFSKS-SIVFGHKVETETRNLIKEITGITKEGGTGLYLGL 117
Query: 350 AETLYAS--INLRFI 388
E S LRF+
Sbjct: 118 PEAFIGSKIETLRFL 132
>ref|NP_654146.1| DUF6, Integral membrane protein DUF6 [Bacillus anthracis A2012]
Length = 309
Score = 32.7 bits (73), Expect = 3.6
Identities = 21/58 (36%), Positives = 32/58 (54%)
Frame = +2
Query: 386 IMLSSTLQSLYPFCWWCRKLVIYQLMFISNFSITSILGTFFG*VFRCATADSSFFMTL 559
+M++S +++L P C L Y+ + + ITS+L F G VF D SFF+TL
Sbjct: 87 VMVASIMEALMPMISICI-LWGYKHVKPKKYMITSMLIAFIGAVFVITKGDMSFFLTL 143
>ref|NP_691788.1| hypothetical protein [Oceanobacillus iheyensis HTE831]
gi|22776548|dbj|BAC12823.1| hypothetical conserved
protein [Oceanobacillus iheyensis HTE831]
Length = 278
Score = 32.7 bits (73), Expect = 3.6
Identities = 17/51 (33%), Positives = 29/51 (56%)
Frame = -3
Query: 571 ETSNEGHEKAGISSSTPENLPEESSQDRGDAKVTDEHELVDDQFSTPPTKR 419
+TSN+ EK S+ TP PE+S + D + D E ++ +TPP+++
Sbjct: 89 DTSNK-EEKQSESNETPAQ-PEQSEEQNNDQQANDSGEQASEEEATPPSQQ 137
>ref|NP_207698.1| hypothetical protein [Helicobacter pylori 26695]
gi|7464410|pir||B64633 hypothetical protein HP0906 -
Helicobacter pylori (strain 26695)
gi|2314048|gb|AAD07958.1| H. pylori predicted coding
region HP0906 [Helicobacter pylori 26695]
Length = 527
Score = 32.3 bits (72), Expect = 4.7
Identities = 20/60 (33%), Positives = 29/60 (48%)
Frame = -3
Query: 592 PSPADNLETSNEGHEKAGISSSTPENLPEESSQDRGDAKVTDEHELVDDQFSTPPTKRVK 413
P P DN+ T E EKA +S E +E+S +A+ T + D ST P + +K
Sbjct: 335 PPPNDNIPTPLEKEEKAKEASDNKEK-TKETSNSAQNAQNTQASDKTSDNKSTAPKETIK 393
>ref|NP_051962.1| gp073R [Rabbit fibroma virus] gi|6578601|gb|AAF17955.1|AF170722_73
gp073R [Rabbit fibroma virus]
Length = 198
Score = 32.0 bits (71), Expect = 6.1
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 1/82 (1%)
Frame = +2
Query: 173 VSELSRIH*EYTTNTWSKTSFTKVYPKLRGKK*KLETKSLIKEVTVIRRKGNSSKLFGEA 352
V E + H E T+NT + T T + KL T+ +IKE+ + K ++
Sbjct: 96 VEESPKSHQESTSNTQNTTPMTDTRDTMDTSDLKLVTEQIIKEMKALNAKVSAVSTI--L 153
Query: 353 ETLY-ASINLRFIMLSSTLQSL 415
ET+ AS+ ++ ML+ ++ L
Sbjct: 154 ETIQAASVGRQYTMLTKNIEEL 175
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 474,907,627
Number of Sequences: 1393205
Number of extensions: 9594429
Number of successful extensions: 24928
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 24293
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24916
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22854740960
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)