Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
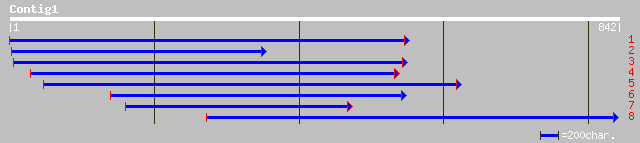
Query= KMC001695A_C01 KMC001695A_c01
(833 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T52269 1,2-diacylglycerol 3-beta-galactosyltransferase (EC ... 249 4e-65
ref|NP_568394.1| monogalactosyldiacylglycerol synthase; protein ... 249 4e-65
ref|NP_565352.1| putative monogalactosyldiacylglycerol synthase;... 229 4e-59
pir||C84499 probable monogalactosyldiacylglycerol synthase [impo... 229 4e-59
pir||T10478 probable 1,2-diacylglycerol 3-beta-galactosyltransfe... 193 2e-48
>pir||T52269 1,2-diacylglycerol 3-beta-galactosyltransferase (EC 2.4.1.46)
[imported] - Arabidopsis thaliana
gi|3367638|emb|CAA04005.1| monogalactosyldiacylglycerol
synthase [Arabidopsis thaliana]
Length = 468
Score = 249 bits (636), Expect = 4e-65
Identities = 121/154 (78%), Positives = 135/154 (87%), Gaps = 1/154 (0%)
Frame = -3
Query: 831 ICGRNKSLASTLESLEWKVPVVIRGFEKQMAKWMGACDCIITKAGPGTIAEALIRGLPIV 652
ICGRNK LAS LE+++WK+PV +RGFE QM KWMGACDCIITKAGPGTIAE+LIR LPI+
Sbjct: 315 ICGRNKKLASALEAIDWKIPVKVRGFETQMEKWMGACDCIITKAGPGTIAESLIRSLPII 374
Query: 651 LNDYIPGQEKGNVPYVVNNGAGVFTRSPKETARIVAEWFSTKSDERKTVAENARKLAQPE 472
LNDYIPGQEKGNVPYVV NGAGVFTRSPKETARIV EWFSTK+DE + ++NARKLAQPE
Sbjct: 375 LNDYIPGQEKGNVPYVVENGAGVFTRSPKETARIVGEWFSTKTDELEQTSDNARKLAQPE 434
Query: 471 AVFDIVRDIHELAEQREP-AHFPYLLTSSFTSLI 373
AVFDIV+DI EL+EQR P A Y LTSSF SL+
Sbjct: 435 AVFDIVKDIDELSEQRGPLASVSYNLTSSFASLV 468
>ref|NP_568394.1| monogalactosyldiacylglycerol synthase; protein id: At5g20410.1,
supported by cDNA: gi_13937144, supported by cDNA:
gi_18700257 [Arabidopsis thaliana]
gi|13937145|gb|AAK50066.1|AF372926_1 AT5g20410/F5O24_300
[Arabidopsis thaliana] gi|18700258|gb|AAL77739.1|
AT5g20410/F5O24_300 [Arabidopsis thaliana]
Length = 394
Score = 249 bits (636), Expect = 4e-65
Identities = 121/154 (78%), Positives = 135/154 (87%), Gaps = 1/154 (0%)
Frame = -3
Query: 831 ICGRNKSLASTLESLEWKVPVVIRGFEKQMAKWMGACDCIITKAGPGTIAEALIRGLPIV 652
ICGRNK LAS LE+++WK+PV +RGFE QM KWMGACDCIITKAGPGTIAE+LIR LPI+
Sbjct: 241 ICGRNKKLASALEAIDWKIPVKVRGFETQMEKWMGACDCIITKAGPGTIAESLIRSLPII 300
Query: 651 LNDYIPGQEKGNVPYVVNNGAGVFTRSPKETARIVAEWFSTKSDERKTVAENARKLAQPE 472
LNDYIPGQEKGNVPYVV NGAGVFTRSPKETARIV EWFSTK+DE + ++NARKLAQPE
Sbjct: 301 LNDYIPGQEKGNVPYVVENGAGVFTRSPKETARIVGEWFSTKTDELEQTSDNARKLAQPE 360
Query: 471 AVFDIVRDIHELAEQREP-AHFPYLLTSSFTSLI 373
AVFDIV+DI EL+EQR P A Y LTSSF SL+
Sbjct: 361 AVFDIVKDIDELSEQRGPLASVSYNLTSSFASLV 394
>ref|NP_565352.1| putative monogalactosyldiacylglycerol synthase; protein id:
At2g11810.1, supported by cDNA: gi_9927294 [Arabidopsis
thaliana] gi|9927295|dbj|BAB12041.1| MGDG synthase type
C [Arabidopsis thaliana] gi|20198177|gb|AAD28678.2|
putative monogalactosyldiacylglycerol synthase
[Arabidopsis thaliana]
Length = 465
Score = 229 bits (584), Expect = 4e-59
Identities = 108/137 (78%), Positives = 123/137 (88%)
Frame = -3
Query: 831 ICGRNKSLASTLESLEWKVPVVIRGFEKQMAKWMGACDCIITKAGPGTIAEALIRGLPIV 652
ICGRNK LASTL S EWK+PV +RGFE QM KWMGACDCIITKAGPGTIAEALI GLPI+
Sbjct: 319 ICGRNKVLASTLASHEWKIPVKVRGFETQMEKWMGACDCIITKAGPGTIAEALICGLPII 378
Query: 651 LNDYIPGQEKGNVPYVVNNGAGVFTRSPKETARIVAEWFSTKSDERKTVAENARKLAQPE 472
LNDYIPGQEKGNVPYVV+NGAGVFTRSPKETA+IVA+WFS +E K ++ENA KL+QPE
Sbjct: 379 LNDYIPGQEKGNVPYVVDNGAGVFTRSPKETAKIVADWFSNNKEELKKMSENALKLSQPE 438
Query: 471 AVFDIVRDIHELAEQRE 421
AVFDIV+DIH L++Q++
Sbjct: 439 AVFDIVKDIHHLSQQQQ 455
>pir||C84499 probable monogalactosyldiacylglycerol synthase [imported] -
Arabidopsis thaliana
Length = 464
Score = 229 bits (584), Expect = 4e-59
Identities = 108/137 (78%), Positives = 123/137 (88%)
Frame = -3
Query: 831 ICGRNKSLASTLESLEWKVPVVIRGFEKQMAKWMGACDCIITKAGPGTIAEALIRGLPIV 652
ICGRNK LASTL S EWK+PV +RGFE QM KWMGACDCIITKAGPGTIAEALI GLPI+
Sbjct: 318 ICGRNKVLASTLASHEWKIPVKVRGFETQMEKWMGACDCIITKAGPGTIAEALICGLPII 377
Query: 651 LNDYIPGQEKGNVPYVVNNGAGVFTRSPKETARIVAEWFSTKSDERKTVAENARKLAQPE 472
LNDYIPGQEKGNVPYVV+NGAGVFTRSPKETA+IVA+WFS +E K ++ENA KL+QPE
Sbjct: 378 LNDYIPGQEKGNVPYVVDNGAGVFTRSPKETAKIVADWFSNNKEELKKMSENALKLSQPE 437
Query: 471 AVFDIVRDIHELAEQRE 421
AVFDIV+DIH L++Q++
Sbjct: 438 AVFDIVKDIHHLSQQQQ 454
>pir||T10478 probable 1,2-diacylglycerol 3-beta-galactosyltransferase (EC
2.4.1.46) precursor, chloroplast - cucumber
gi|1805254|gb|AAC49624.1| monogalactosyldiacylglycerol
synthase [Cucumis sativus]
Length = 525
Score = 193 bits (491), Expect = 2e-48
Identities = 90/136 (66%), Positives = 110/136 (80%)
Frame = -3
Query: 831 ICGRNKSLASTLESLEWKVPVVIRGFEKQMAKWMGACDCIITKAGPGTIAEALIRGLPIV 652
ICG NK LA L S++WKVPV ++GF +M + MGACDCIITKAGPGTIAEA+IRGLPI+
Sbjct: 382 ICGHNKKLAGRLRSIDWKVPVQVKGFVTKMEECMGACDCIITKAGPGTIAEAMIRGLPII 441
Query: 651 LNDYIPGQEKGNVPYVVNNGAGVFTRSPKETARIVAEWFSTKSDERKTVAENARKLAQPE 472
LNDYI GQE GNVPYVV NG G F++SPKE A IVA+WF K+DE +++NA +LA+P+
Sbjct: 442 LNDYIAGQEAGNVPYVVENGCGKFSKSPKEIANIVAKWFGPKADELLIMSQNALRLARPD 501
Query: 471 AVFDIVRDIHELAEQR 424
AVF IV D+HEL +QR
Sbjct: 502 AVFKIVHDLHELVKQR 517
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 660,713,842
Number of Sequences: 1393205
Number of extensions: 13743301
Number of successful extensions: 34461
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 33715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34450
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 43201326735
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)