Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001682A_C01 KMC001682A_c01
(562 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
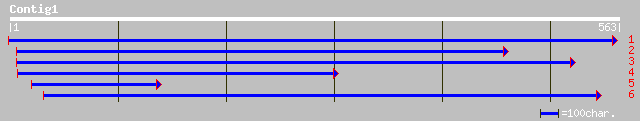
gb|AAM62971.1| putative xyloglucan endotransglycosylase [Arabido... 199 3e-50
ref|NP_192230.1| xyloglucan endotransglycosylase, putative; prot... 199 3e-50
gb|AAC09388.1| xyloglucan endotransglycosylase precursor [Actini... 135 3e-31
pir||B49539 xyloglucan endo-1,4-beta-D-glucanase (EC 3.2.1.-) - ... 135 3e-31
dbj|BAA03922.1| endo-xyloglucan transferase [Glycine max] 135 3e-31
>gb|AAM62971.1| putative xyloglucan endotransglycosylase [Arabidopsis thaliana]
Length = 287
Score = 199 bits (505), Expect = 3e-50
Identities = 85/128 (66%), Positives = 105/128 (81%)
Frame = -1
Query: 556 NMEHRGIPFPKDQPMGVYSSIWNADDWATQGGRVKTDWSHAPFIATYKSFEINACECPVS 377
N+E +GIPF KDQ MGVYSSIWNADDWATQGG VKTDWSHAPF+A+YK F+I+ACE P +
Sbjct: 166 NLEEKGIPFAKDQAMGVYSSIWNADDWATQGGLVKTDWSHAPFVASYKEFQIDACEIPTT 225
Query: 376 SVAAVDNAKRCSSSEEKKFWWDEPTLSELSLHQSHQLMWVKARHMVYDYCTDISRYPVTP 197
+ + +C+ ++KFWWDEPT+SELSLHQ+HQL+WV+A HM+YDYC D +R+PVTP
Sbjct: 226 T-----DLSKCNG--DQKFWWDEPTVSELSLHQNHQLIWVRANHMIYDYCFDATRFPVTP 278
Query: 196 AECVHHHH 173
EC HH H
Sbjct: 279 LECQHHRH 286
>ref|NP_192230.1| xyloglucan endotransglycosylase, putative; protein id: At4g03210.1,
supported by cDNA: 17748., supported by cDNA:
gi_14994278, supported by cDNA: gi_18252836 [Arabidopsis
thaliana] gi|25313540|pir||G85040 probable xyloglucan
endotransglycosylase [imported] - Arabidopsis thaliana
gi|4262149|gb|AAD14449.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|7270191|emb|CAB77806.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|14994279|gb|AAK73274.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|18252837|gb|AAL62345.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|25084277|gb|AAN72210.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
Length = 290
Score = 199 bits (505), Expect = 3e-50
Identities = 85/128 (66%), Positives = 105/128 (81%)
Frame = -1
Query: 556 NMEHRGIPFPKDQPMGVYSSIWNADDWATQGGRVKTDWSHAPFIATYKSFEINACECPVS 377
N+E +GIPF KDQ MGVYSSIWNADDWATQGG VKTDWSHAPF+A+YK F+I+ACE P +
Sbjct: 169 NLEEKGIPFAKDQAMGVYSSIWNADDWATQGGLVKTDWSHAPFVASYKEFQIDACEIPTT 228
Query: 376 SVAAVDNAKRCSSSEEKKFWWDEPTLSELSLHQSHQLMWVKARHMVYDYCTDISRYPVTP 197
+ + +C+ ++KFWWDEPT+SELSLHQ+HQL+WV+A HM+YDYC D +R+PVTP
Sbjct: 229 T-----DLSKCNG--DQKFWWDEPTVSELSLHQNHQLIWVRANHMIYDYCFDATRFPVTP 281
Query: 196 AECVHHHH 173
EC HH H
Sbjct: 282 LECQHHRH 289
>gb|AAC09388.1| xyloglucan endotransglycosylase precursor [Actinidia deliciosa]
Length = 293
Score = 135 bits (341), Expect = 3e-31
Identities = 61/123 (49%), Positives = 81/123 (65%)
Frame = -1
Query: 556 NMEHRGIPFPKDQPMGVYSSIWNADDWATQGGRVKTDWSHAPFIATYKSFEINACECPVS 377
N + G+ FP DQPM +YSS+WNADDWAT+GG KTDWS APF+A YKSF I+ CE V
Sbjct: 174 NSKDLGVKFPFDQPMKIYSSLWNADDWATRGGLEKTDWSKAPFVAAYKSFHIDGCEASV- 232
Query: 376 SVAAVDNAKRCSSSEEKKFWWDEPTLSELSLHQSHQLMWVKARHMVYDYCTDISRYPVTP 197
AK C++ + K WWD+ +L Q +L WV++++ +Y+YCTD +RYP P
Sbjct: 233 ------EAKFCAT--QGKRWWDQNDYRDLDAFQYRRLRWVRSKYTIYNYCTDRTRYPTMP 284
Query: 196 AEC 188
EC
Sbjct: 285 PEC 287
>pir||B49539 xyloglucan endo-1,4-beta-D-glucanase (EC 3.2.1.-) - soybean
Length = 292
Score = 135 bits (340), Expect = 3e-31
Identities = 62/124 (50%), Positives = 81/124 (65%), Gaps = 1/124 (0%)
Frame = -1
Query: 556 NMEHRGIPFPKDQPMGVYSSIWNADDWATQGGRVKTDWSHAPFIATYKSFEINACECPVS 377
N + G+ FP DQPM +Y+S+WNADDWAT+GG KTDWS APFIA YK F I+ CE V
Sbjct: 172 NSKDLGVKFPFDQPMKIYNSLWNADDWATRGGLEKTDWSKAPFIAAYKGFHIDGCEASV- 230
Query: 376 SVAAVDNAKRCSSSEEKKFWWDEPTLSELSLHQSHQLMWVKARHMVYDYCTDISRYP-VT 200
NAK C + + K WWD+P +L Q +L WV+ ++ +Y+YCTD RYP ++
Sbjct: 231 ------NAKFCDT--QGKRWWDQPEFRDLDAAQWRRLRWVRQKYTIYNYCTDTKRYPHIS 282
Query: 199 PAEC 188
P EC
Sbjct: 283 PPEC 286
>dbj|BAA03922.1| endo-xyloglucan transferase [Glycine max]
Length = 295
Score = 135 bits (340), Expect = 3e-31
Identities = 62/124 (50%), Positives = 81/124 (65%), Gaps = 1/124 (0%)
Frame = -1
Query: 556 NMEHRGIPFPKDQPMGVYSSIWNADDWATQGGRVKTDWSHAPFIATYKSFEINACECPVS 377
N + G+ FP DQPM +Y+S+WNADDWAT+GG KTDWS APFIA YK F I+ CE V
Sbjct: 175 NSKDLGVKFPFDQPMKIYNSLWNADDWATRGGLEKTDWSKAPFIAAYKGFHIDGCEASV- 233
Query: 376 SVAAVDNAKRCSSSEEKKFWWDEPTLSELSLHQSHQLMWVKARHMVYDYCTDISRYP-VT 200
NAK C + + K WWD+P +L Q +L WV+ ++ +Y+YCTD RYP ++
Sbjct: 234 ------NAKFCDT--QGKRWWDQPEFRDLDAAQWRRLRWVRQKYTIYNYCTDTKRYPHIS 285
Query: 199 PAEC 188
P EC
Sbjct: 286 PPEC 289
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,823,420
Number of Sequences: 1393205
Number of extensions: 12768150
Number of successful extensions: 35508
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 33976
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35362
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)