Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001661A_C01 KMC001661A_c01
(659 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
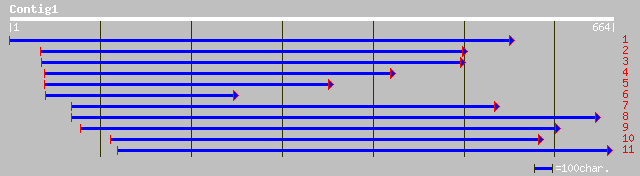
gb|AAK63930.1|AC084282_11 putative dehydrogenase precursor [Oryz... 232 4e-60
ref|NP_172934.1| aspartate-semialdehyde dehydrogenase, putative;... 230 1e-59
gb|AAG33078.1|AF207064_1 aspartate-semialdehyde dehydrogenase pr... 230 1e-59
pir||B86282 protein F10B6.22 [imported] - Arabidopsis thaliana g... 214 1e-54
gb|ZP_00107922.1| hypothetical protein [Nostoc punctiforme] 162 4e-39
>gb|AAK63930.1|AC084282_11 putative dehydrogenase precursor [Oryza sativa (japonica
cultivar-group)]
Length = 375
Score = 232 bits (591), Expect = 4e-60
Identities = 114/128 (89%), Positives = 122/128 (95%)
Frame = -2
Query: 658 YNEEEMKMVKETRKIWNDKDVKVTATCIRVPVMRAHAESVNLQFERPLDEDTAREILKNA 479
YNEEEMKMVKETRKIWNDKDVKVTATCIRVPVMRAHAESVNLQFE+PLDEDTAREIL+ A
Sbjct: 247 YNEEEMKMVKETRKIWNDKDVKVTATCIRVPVMRAHAESVNLQFEKPLDEDTAREILRAA 306
Query: 478 PGVVVIDDREANHFPTPLEVSNKDDVAVGRIRRDLSQDGNQGLDIFVCGDQIRKGAALNA 299
GV +IDDR +N FPTPLEVS+KDDVAVGRIR+DLSQD N+GLDIFVCGDQIRKGAALNA
Sbjct: 307 EGVTIIDDRASNRFPTPLEVSDKDDVAVGRIRQDLSQDDNKGLDIFVCGDQIRKGAALNA 366
Query: 298 IQIAEMLL 275
+QIAEMLL
Sbjct: 367 VQIAEMLL 374
>ref|NP_172934.1| aspartate-semialdehyde dehydrogenase, putative; protein id:
At1g14810.1, supported by cDNA: 109497., supported by
cDNA: gi_17979523, supported by cDNA: gi_20856223
[Arabidopsis thaliana] gi|17979524|gb|AAL50097.1|
At1g14810/F10B6_6 [Arabidopsis thaliana]
gi|20856224|gb|AAM26654.1| At1g14810/F10B6_6
[Arabidopsis thaliana] gi|21536731|gb|AAM61063.1|
aspartate-semialdehyde dehydrogenase, putative
[Arabidopsis thaliana]
Length = 375
Score = 230 bits (587), Expect = 1e-59
Identities = 113/128 (88%), Positives = 121/128 (94%)
Frame = -2
Query: 658 YNEEEMKMVKETRKIWNDKDVKVTATCIRVPVMRAHAESVNLQFERPLDEDTAREILKNA 479
YNEEEMK+VKETRKIWND +VKVTATCIRVPVMRAHAESVNLQFE PLDE+TAREILK A
Sbjct: 248 YNEEEMKLVKETRKIWNDTEVKVTATCIRVPVMRAHAESVNLQFENPLDENTAREILKKA 307
Query: 478 PGVVVIDDREANHFPTPLEVSNKDDVAVGRIRRDLSQDGNQGLDIFVCGDQIRKGAALNA 299
PGV +IDDR +N FPTPL+VSNKDDVAVGRIRRD+SQDGN GLDIFVCGDQIRKGAALNA
Sbjct: 308 PGVYIIDDRASNTFPTPLDVSNKDDVAVGRIRRDVSQDGNFGLDIFVCGDQIRKGAALNA 367
Query: 298 IQIAEMLL 275
+QIAEMLL
Sbjct: 368 VQIAEMLL 375
>gb|AAG33078.1|AF207064_1 aspartate-semialdehyde dehydrogenase precursor [Arabidopsis
thaliana]
Length = 340
Score = 230 bits (587), Expect = 1e-59
Identities = 113/128 (88%), Positives = 121/128 (94%)
Frame = -2
Query: 658 YNEEEMKMVKETRKIWNDKDVKVTATCIRVPVMRAHAESVNLQFERPLDEDTAREILKNA 479
YNEEEMK+VKETRKIWND +VKVTATCIRVPVMRAHAESVNLQFE PLDE+TAREILK A
Sbjct: 213 YNEEEMKLVKETRKIWNDTEVKVTATCIRVPVMRAHAESVNLQFENPLDENTAREILKKA 272
Query: 478 PGVVVIDDREANHFPTPLEVSNKDDVAVGRIRRDLSQDGNQGLDIFVCGDQIRKGAALNA 299
PGV +IDDR +N FPTPL+VSNKDDVAVGRIRRD+SQDGN GLDIFVCGDQIRKGAALNA
Sbjct: 273 PGVYIIDDRASNTFPTPLDVSNKDDVAVGRIRRDVSQDGNFGLDIFVCGDQIRKGAALNA 332
Query: 298 IQIAEMLL 275
+QIAEMLL
Sbjct: 333 VQIAEMLL 340
>pir||B86282 protein F10B6.22 [imported] - Arabidopsis thaliana
gi|8778230|gb|AAF79239.1|AC006917_24 F10B6.22
[Arabidopsis thaliana]
Length = 730
Score = 214 bits (544), Expect = 1e-54
Identities = 113/160 (70%), Positives = 121/160 (75%), Gaps = 32/160 (20%)
Frame = -2
Query: 658 YNEEEMKMVKETRKIW--------------------------------NDKDVKVTATCI 575
YNEEEMK+VKETRKIW ND +VKVTATCI
Sbjct: 571 YNEEEMKLVKETRKIWVSGYLSAILCSFHFDLLFKNFRYLTPISCPSQNDTEVKVTATCI 630
Query: 574 RVPVMRAHAESVNLQFERPLDEDTAREILKNAPGVVVIDDREANHFPTPLEVSNKDDVAV 395
RVPVMRAHAESVNLQFE PLDE+TAREILK APGV +IDDR +N FPTPL+VSNKDDVAV
Sbjct: 631 RVPVMRAHAESVNLQFENPLDENTAREILKKAPGVYIIDDRASNTFPTPLDVSNKDDVAV 690
Query: 394 GRIRRDLSQDGNQGLDIFVCGDQIRKGAALNAIQIAEMLL 275
GRIRRD+SQDGN GLDIFVCGDQIRKGAALNA+QIAEMLL
Sbjct: 691 GRIRRDVSQDGNFGLDIFVCGDQIRKGAALNAVQIAEMLL 730
>gb|ZP_00107922.1| hypothetical protein [Nostoc punctiforme]
Length = 287
Score = 162 bits (410), Expect = 4e-39
Identities = 76/128 (59%), Positives = 105/128 (81%)
Frame = -2
Query: 658 YNEEEMKMVKETRKIWNDKDVKVTATCIRVPVMRAHAESVNLQFERPLDEDTAREILKNA 479
Y EEEMKMV ETRKI+ + +++TATCIRVPV+RAH+E++NL+FE + AREIL ++
Sbjct: 150 YCEEEMKMVNETRKIFGTQQIRITATCIRVPVLRAHSEAINLEFETTFSAEEAREILNSS 209
Query: 478 PGVVVIDDREANHFPTPLEVSNKDDVAVGRIRRDLSQDGNQGLDIFVCGDQIRKGAALNA 299
PGV +++D E NHFP P+E + +D+V VGRIR+D+S + GL++++CGDQIRKGAALNA
Sbjct: 210 PGVKLVEDWETNHFPMPIEATGRDEVLVGRIRQDISH--SCGLELWLCGDQIRKGAALNA 267
Query: 298 IQIAEMLL 275
IQIAE+L+
Sbjct: 268 IQIAELLV 275
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 540,694,429
Number of Sequences: 1393205
Number of extensions: 11381666
Number of successful extensions: 48159
Number of sequences better than 10.0: 238
Number of HSP's better than 10.0 without gapping: 46675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 47944
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28289785200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)