Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001646A_C01 KMC001646A_c01
(612 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
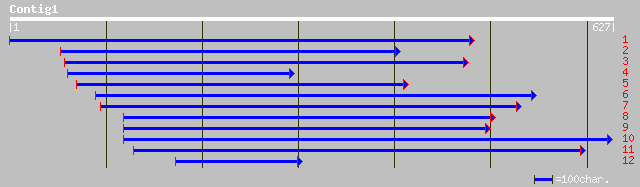
gb|AAL16064.1|AF420238_1 S-adenosyl-L-methionine synthetase [Den... 119 4e-42
gb|AAG42490.1| S-adenosylmethionine sythetase 2 [Suaeda maritima... 116 8e-42
sp|P50300|METK_PINBN S-adenosylmethionine synthetase (Methionine... 115 2e-41
sp|P93438|METL_ORYSA S-adenosylmethionine synthetase 2 (Methioni... 117 7e-41
sp|P50299|METK_HORVU S-adenosylmethionine synthetase 1 (Methioni... 116 9e-41
>gb|AAL16064.1|AF420238_1 S-adenosyl-L-methionine synthetase [Dendrobium crumenatum]
Length = 395
Score = 119 bits (298), Expect(2) = 4e-42
Identities = 58/66 (87%), Positives = 62/66 (93%)
Frame = -3
Query: 610 SIVAVGLARRCLVQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKEEFDFRPGMISIN 431
S+VA GLARRC+VQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKE FDFRPGMI+IN
Sbjct: 295 SVVANGLARRCIVQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKENFDFRPGMITIN 354
Query: 430 LDPQEG 413
LD + G
Sbjct: 355 LDLKRG 360
Score = 74.3 bits (181), Expect(2) = 4e-42
Identities = 34/39 (87%), Positives = 36/39 (92%)
Frame = -1
Query: 429 LILKRGGNNRFLKTAAYGHFGRDDADFTWEVVKPLKWEK 313
L LKRGGN RF+KTAAYGHFGRDD DFTWEVVKPLKW+K
Sbjct: 355 LDLKRGGN-RFIKTAAYGHFGRDDPDFTWEVVKPLKWDK 392
>gb|AAG42490.1| S-adenosylmethionine sythetase 2 [Suaeda maritima subsp. salsa]
Length = 395
Score = 116 bits (290), Expect(2) = 8e-42
Identities = 58/66 (87%), Positives = 61/66 (91%)
Frame = -3
Query: 610 SIVAVGLARRCLVQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKEEFDFRPGMISIN 431
SIVA GLARR +VQVSYAIGVPEPLSVFVDTYGTGKIPDK+ILKIVKE FDFRPGMISIN
Sbjct: 292 SIVANGLARRAIVQVSYAIGVPEPLSVFVDTYGTGKIPDKDILKIVKENFDFRPGMISIN 351
Query: 430 LDPQEG 413
LD + G
Sbjct: 352 LDLKRG 357
Score = 76.3 bits (186), Expect(2) = 8e-42
Identities = 34/39 (87%), Positives = 34/39 (87%)
Frame = -1
Query: 429 LILKRGGNNRFLKTAAYGHFGRDDADFTWEVVKPLKWEK 313
L LKRGGN RF KTAAYGHFGRDD DFTWE VKPLKWEK
Sbjct: 352 LDLKRGGNARFQKTAAYGHFGRDDPDFTWETVKPLKWEK 390
>sp|P50300|METK_PINBN S-adenosylmethionine synthetase (Methionine adenosyltransferase)
(AdoMet synthetase) gi|1033190|gb|AAA79831.1| S-adenosyl
methionine synthetase
Length = 393
Score = 115 bits (289), Expect(2) = 2e-41
Identities = 56/66 (84%), Positives = 61/66 (91%)
Frame = -3
Query: 610 SIVAVGLARRCLVQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKEEFDFRPGMISIN 431
SIVA GLARRC+VQVSYAIGVPEPLS+FVDTYGTG IPDKEIL+I+KE FDFRPGMISIN
Sbjct: 292 SIVANGLARRCIVQVSYAIGVPEPLSLFVDTYGTGSIPDKEILEIIKEHFDFRPGMISIN 351
Query: 430 LDPQEG 413
LD + G
Sbjct: 352 LDLKRG 357
Score = 75.5 bits (184), Expect(2) = 2e-41
Identities = 34/40 (85%), Positives = 34/40 (85%)
Frame = -1
Query: 429 LILKRGGNNRFLKTAAYGHFGRDDADFTWEVVKPLKWEKA 310
L LKRGGN R KTAAYGHFGRDD DFTWE VKPLKWEKA
Sbjct: 352 LDLKRGGNGRSQKTAAYGHFGRDDPDFTWETVKPLKWEKA 391
>sp|P93438|METL_ORYSA S-adenosylmethionine synthetase 2 (Methionine adenosyltransferase
2) (AdoMet synthetase 2) gi|1778821|gb|AAC05590.1|
S-adenosyl-L-methionine synthetase [Oryza sativa]
gi|29027806|dbj|BAC65881.1| OSJNBa0011P19.5 [Oryza
sativa (japonica cultivar-group)]
Length = 394
Score = 117 bits (293), Expect(2) = 7e-41
Identities = 57/66 (86%), Positives = 62/66 (93%)
Frame = -3
Query: 610 SIVAVGLARRCLVQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKEEFDFRPGMISIN 431
SIVA GLARRC+VQVSYAIGVPEPLSVFVD+YGTGKIPDKEILKIVKE FDFRPGM++IN
Sbjct: 294 SIVASGLARRCIVQVSYAIGVPEPLSVFVDSYGTGKIPDKEILKIVKENFDFRPGMMTIN 353
Query: 430 LDPQEG 413
LD + G
Sbjct: 354 LDLKRG 359
Score = 72.0 bits (175), Expect(2) = 7e-41
Identities = 34/40 (85%), Positives = 37/40 (92%)
Frame = -1
Query: 429 LILKRGGNNRFLKTAAYGHFGRDDADFTWEVVKPLKWEKA 310
L LKRGGN RF+KTAAYGHFGR+D DFTWEVVKPLK+EKA
Sbjct: 354 LDLKRGGN-RFIKTAAYGHFGREDPDFTWEVVKPLKYEKA 392
>sp|P50299|METK_HORVU S-adenosylmethionine synthetase 1 (Methionine adenosyltransferase
1) (AdoMet synthetase 1) gi|7434000|pir||T06180
methionine adenosyltransferase (EC 2.5.1.6) - barley
gi|960357|dbj|BAA09895.1| S-adenosylmethionine
synthetase [Hordeum vulgare]
Length = 394
Score = 116 bits (290), Expect(2) = 9e-41
Identities = 54/66 (81%), Positives = 63/66 (94%)
Frame = -3
Query: 610 SIVAVGLARRCLVQVSYAIGVPEPLSVFVDTYGTGKIPDKEILKIVKEEFDFRPGMISIN 431
SI+A GLARRC+VQ+SYAIGVPEPLSVFVD+YGTGKIPD+EILK+VKE FDFRPGMI+IN
Sbjct: 294 SIIASGLARRCIVQISYAIGVPEPLSVFVDSYGTGKIPDREILKLVKENFDFRPGMITIN 353
Query: 430 LDPQEG 413
LD ++G
Sbjct: 354 LDLKKG 359
Score = 72.8 bits (177), Expect(2) = 9e-41
Identities = 34/40 (85%), Positives = 38/40 (95%)
Frame = -1
Query: 429 LILKRGGNNRFLKTAAYGHFGRDDADFTWEVVKPLKWEKA 310
L LK+GGN RF+KTAAYGHFGRDDADFTWEVVKPLK++KA
Sbjct: 354 LDLKKGGN-RFIKTAAYGHFGRDDADFTWEVVKPLKFDKA 392
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 530,345,525
Number of Sequences: 1393205
Number of extensions: 11530011
Number of successful extensions: 36842
Number of sequences better than 10.0: 263
Number of HSP's better than 10.0 without gapping: 34163
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36603
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)