Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001643A_C01 KMC001643A_c01
(635 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
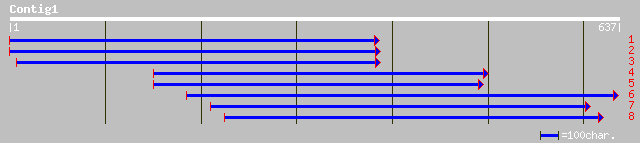
emb|CAC35729.1| membrane transporter [Lotus japonicus] 181 7e-45
pir||T06237 probable high affinity nitrate transporter protein -... 160 2e-38
pir||T16972 probable high affinity nitrate transporter protein -... 142 4e-33
gb|AAF00054.1|AF092654_1 putative high-affinity nitrate transpor... 141 8e-33
gb|AAF00053.1|AF092655_1 putative high-affinity nitrate transpor... 141 8e-33
>emb|CAC35729.1| membrane transporter [Lotus japonicus]
Length = 530
Score = 181 bits (459), Expect = 7e-45
Identities = 84/86 (97%), Positives = 86/86 (99%)
Frame = -2
Query: 634 MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDIHKSSEEHYYTAEWDEEERKKGLHSQSLKF 455
MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDI+KSSEEHYYTAEWDEEERKKGLHSQSLKF
Sbjct: 445 MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDINKSSEEHYYTAEWDEEERKKGLHSQSLKF 504
Query: 454 AENSRSERGKRISSAPTPPNTTPTHV 377
AENSRSERGKR+SSAPTPPNTTPTHV
Sbjct: 505 AENSRSERGKRVSSAPTPPNTTPTHV 530
>pir||T06237 probable high affinity nitrate transporter protein - soybean
gi|3005576|gb|AAC09320.1| putative high affinity nitrate
transporter; GmNRT2 [Glycine max]
Length = 530
Score = 160 bits (404), Expect = 2e-38
Identities = 71/86 (82%), Positives = 81/86 (93%)
Frame = -2
Query: 634 MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDIHKSSEEHYYTAEWDEEERKKGLHSQSLKF 455
MGVMIV CTLPV++VHFPQWGSMFLPPSKD+ KS+EE YYT+EW+EEE++KGLH QSLKF
Sbjct: 445 MGVMIVACTLPVSVVHFPQWGSMFLPPSKDVSKSTEEFYYTSEWNEEEKQKGLHQQSLKF 504
Query: 454 AENSRSERGKRISSAPTPPNTTPTHV 377
AENSRSERGKR++SAPTPPN TPTHV
Sbjct: 505 AENSRSERGKRVASAPTPPNATPTHV 530
>pir||T16972 probable high affinity nitrate transporter protein - curled-leaved
tobacco gi|2208960|emb|CAA69387.1| nitrate transporter
[Nicotiana plumbaginifolia]
Length = 530
Score = 142 bits (358), Expect = 4e-33
Identities = 64/86 (74%), Positives = 71/86 (82%)
Frame = -2
Query: 634 MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDIHKSSEEHYYTAEWDEEERKKGLHSQSLKF 455
MG+MI+GCTLPVTL HFPQWGSMF PP+KD K SEEHYY AE+ E ER+KG+H SLKF
Sbjct: 445 MGMMIIGCTLPVTLCHFPQWGSMFFPPTKDPVKGSEEHYYAAEYTEAERQKGMHQNSLKF 504
Query: 454 AENSRSERGKRISSAPTPPNTTPTHV 377
AEN RSERGKR+ SAPTPPN TP V
Sbjct: 505 AENCRSERGKRVGSAPTPPNLTPNRV 530
>gb|AAF00054.1|AF092654_1 putative high-affinity nitrate transporter [Lycopersicon
esculentum]
Length = 530
Score = 141 bits (355), Expect = 8e-33
Identities = 63/86 (73%), Positives = 71/86 (82%)
Frame = -2
Query: 634 MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDIHKSSEEHYYTAEWDEEERKKGLHSQSLKF 455
MG MI+GCTLPVT HFPQWGSMFLPP+KD K +EEHYYT+E+ E ER+KG+H SLKF
Sbjct: 445 MGFMIIGCTLPVTFCHFPQWGSMFLPPTKDPVKGTEEHYYTSEYTEAERQKGMHQNSLKF 504
Query: 454 AENSRSERGKRISSAPTPPNTTPTHV 377
AEN RSERGKR+ SAPTPPN TP V
Sbjct: 505 AENCRSERGKRVGSAPTPPNLTPNRV 530
>gb|AAF00053.1|AF092655_1 putative high-affinity nitrate transporter [Lycopersicon
esculentum]
Length = 530
Score = 141 bits (355), Expect = 8e-33
Identities = 63/86 (73%), Positives = 71/86 (82%)
Frame = -2
Query: 634 MGVMIVGCTLPVTLVHFPQWGSMFLPPSKDIHKSSEEHYYTAEWDEEERKKGLHSQSLKF 455
MG MI+GCTLPVT HFPQWGSMFLPP+KD K +EEHYYT+E+ E ER+KG+H SLKF
Sbjct: 445 MGFMIIGCTLPVTFCHFPQWGSMFLPPTKDPVKGTEEHYYTSEYTEAERQKGMHQNSLKF 504
Query: 454 AENSRSERGKRISSAPTPPNTTPTHV 377
AEN RSERGKR+ SAPTPPN TP V
Sbjct: 505 AENCRSERGKRVGSAPTPPNLTPNRV 530
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 574,759,323
Number of Sequences: 1393205
Number of extensions: 12492782
Number of successful extensions: 35022
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 33508
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34977
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)