Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001616A_C01 KMC001616A_c01
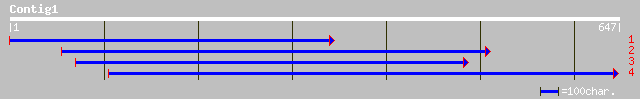
(645 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO18639.1| galactose dehydrogenase [Actinidia deliciosa] 252 3e-66
ref|NP_195093.1| putative protein; protein id: At4g33670.1, supp... 234 7e-61
ref|NP_812525.1| putative oxidoreductase [Bacteroides thetaiotao... 125 6e-28
gb|EAA05276.1| ebiP8370 [Anopheles gambiae str. PEST] 100 1e-20
ref|NP_498580.1| Aldo keto reductase family [Caenorhabditis eleg... 92 8e-18
>gb|AAO18639.1| galactose dehydrogenase [Actinidia deliciosa]
Length = 319
Score = 252 bits (644), Expect = 3e-66
Identities = 121/150 (80%), Positives = 138/150 (91%)
Frame = -3
Query: 643 DRVPPGXLDVVLSYCHHCINDTTLEDLVPYLKSKGVGVINASPLAMGLLTEAGPPEWHPA 464
DRVPPG +DV+LSYCH+ IND+TLEDL+PYLKSKGVGVI+ASPLAMGLLTE+GPPEWHPA
Sbjct: 169 DRVPPGTVDVILSYCHYSINDSTLEDLLPYLKSKGVGVISASPLAMGLLTESGPPEWHPA 228
Query: 463 SAELKSACQAAATHCKEKGKNISKLALQYSLVNKDITSVLVGMRSVEQVEENVAAARELA 284
S ELK+ACQAAA HCKEKG+NISKLA+QYSL NKDI+S+LVGM SV+QVEENVAAA ELA
Sbjct: 229 SPELKAACQAAAAHCKEKGRNISKLAMQYSLSNKDISSILVGMNSVKQVEENVAAANELA 288
Query: 283 ASGIDEETLSEVENILKPVKNQSWPSGIQQ 194
G DE+T+SE+E ILKPVKNQ+W SGIQQ
Sbjct: 289 TFGKDEKTVSEIEEILKPVKNQTWLSGIQQ 318
>ref|NP_195093.1| putative protein; protein id: At4g33670.1, supported by cDNA:
gi_15215697, supported by cDNA: gi_19699263 [Arabidopsis
thaliana] gi|7447943|pir||T04984 hypothetical protein
T16L1.160 - Arabidopsis thaliana
gi|3549669|emb|CAA20580.1| putative protein [Arabidopsis
thaliana] gi|7270315|emb|CAB80084.1| putative protein
[Arabidopsis thaliana] gi|15215698|gb|AAK91395.1|
AT4g33670/T16L1_160 [Arabidopsis thaliana]
gi|16555790|emb|CAD10386.1| L-galactose dehydrogenase
[Arabidopsis thaliana] gi|19699264|gb|AAL90998.1|
AT4g33670/T16L1_160 [Arabidopsis thaliana]
Length = 319
Score = 234 bits (597), Expect = 7e-61
Identities = 114/151 (75%), Positives = 131/151 (86%)
Frame = -3
Query: 643 DRVPPGXLDVVLSYCHHCINDTTLEDLVPYLKSKGVGVINASPLAMGLLTEAGPPEWHPA 464
DRVPPG +DV+LSYCH+ +ND+TL DL+PYLKSKGVGVI+ASPLAMGLLTE GPPEWHPA
Sbjct: 169 DRVPPGTVDVILSYCHYGVNDSTLLDLLPYLKSKGVGVISASPLAMGLLTEQGPPEWHPA 228
Query: 463 SAELKSACQAAATHCKEKGKNISKLALQYSLVNKDITSVLVGMRSVEQVEENVAAARELA 284
S ELKSA +AA HCK KGK I+KLALQYSL NK+I+SVLVGM SV QVEENVAA EL
Sbjct: 229 SPELKSASKAAVAHCKSKGKKITKLALQYSLANKEISSVLVGMSSVSQVEENVAAVTELE 288
Query: 283 ASGIDEETLSEVENILKPVKNQSWPSGIQQS 191
+ G+D+ETLSEVE IL+PVKN +WPSGI Q+
Sbjct: 289 SLGMDQETLSEVEAILEPVKNLTWPSGIHQN 319
>ref|NP_812525.1| putative oxidoreductase [Bacteroides thetaiotaomicron VPI-5482]
gi|29340929|gb|AAO78719.1| putative oxidoreductase
[Bacteroides thetaiotaomicron VPI-5482]
Length = 310
Score = 125 bits (313), Expect = 6e-28
Identities = 63/144 (43%), Positives = 86/144 (58%)
Frame = -3
Query: 643 DRVPPGXLDVVLSYCHHCINDTTLEDLVPYLKSKGVGVINASPLAMGLLTEAGPPEWHPA 464
DR P G ++ VLS+CH+C+ D L D + Y +SK +GVINASPL+MGLL+E G P WHPA
Sbjct: 168 DRSPSGTIESVLSFCHYCLCDDKLADFLDYFESKEIGVINASPLSMGLLSERGVPVWHPA 227
Query: 463 SAELKSACQAAATHCKEKGKNISKLALQYSLVNKDITSVLVGMRSVEQVEENVAAARELA 284
L AC+ A HCK K I KLA+Q+S+ N I + L + E V++N+ E
Sbjct: 228 PKPLVDACRKAMEHCKAKNYPIEKLAMQFSVSNPKIATTLFSTTNPENVKKNIGFIEE-- 285
Query: 283 ASGIDEETLSEVENILKPVKNQSW 212
+D E + EV I+ + SW
Sbjct: 286 --PVDWELVREVREIIGEQQRVSW 307
>gb|EAA05276.1| ebiP8370 [Anopheles gambiae str. PEST]
Length = 342
Score = 100 bits (250), Expect = 1e-20
Identities = 53/141 (37%), Positives = 81/141 (56%), Gaps = 1/141 (0%)
Frame = -3
Query: 631 PGXLDVVLSYCHHCINDTTLEDLVPYLKSKGVGVINASPLAMGLLTEAGPPEWHPASAEL 452
PG D VLSYC + + D +LE+ +P+ + +G+I AS MGLLT GP WHPA +L
Sbjct: 186 PGRFDTVLSYCRNTLFDDSLEEYIPFFREHKMGLICASGHGMGLLTNGGPQPWHPADRQL 245
Query: 451 KSACQAAATHCKEKGKNISKLALQYSLVNKDITSVLVGMRSVEQVEENVAAARELAASGI 272
+ C AA +CK +G + KLA+ ++L + L GM++ E V+ N+ A E S
Sbjct: 246 REVCAEAAAYCKREGVELGKLAMHHALQMPGPATFLAGMQTPELVQINLDAYFE-GLSEK 304
Query: 271 DEETLSEV-ENILKPVKNQSW 212
+ + LS + E + V+N+ W
Sbjct: 305 EADVLSYLKERVFPKVENKHW 325
>ref|NP_498580.1| Aldo keto reductase family [Caenorhabditis elegans]
gi|7500712|pir||T28841 hypothetical protein F37C12.12 -
Caenorhabditis elegans gi|458980|gb|AAC48300.1|
Hypothetical protein F37C12.12 [Caenorhabditis elegans]
Length = 439
Score = 91.7 bits (226), Expect = 8e-18
Identities = 46/113 (40%), Positives = 67/113 (58%), Gaps = 1/113 (0%)
Frame = -3
Query: 547 SKGVGVINASPLAMGLLTEAGPPEWHPASAELKSACQAAATHCKEKGKNISKLALQYSLV 368
++ + VIN+ L GLLTE GPP WHPAS E+K AC AA T+C K +ISKLAL Y+L
Sbjct: 269 TRNIAVINSGALCWGLLTEKGPPPWHPASDEIKEACLAATTYCSSKNISISKLALDYALN 328
Query: 367 NKDITSVLVGMRSVEQVEENVAAARELAASGIDEETLSEV-ENILKPVKNQSW 212
++ LVGM SV+QV +N+ + + +++ + L ++N W
Sbjct: 329 FPNVICCLVGMDSVQQVLDNLELSNFSRITDVEQRVRDRIMRRYLDRLENAGW 381
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 543,934,538
Number of Sequences: 1393205
Number of extensions: 11866596
Number of successful extensions: 37632
Number of sequences better than 10.0: 293
Number of HSP's better than 10.0 without gapping: 35788
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37440
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27291941472
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)