Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001610A_C01 KMC001610A_c01
(637 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
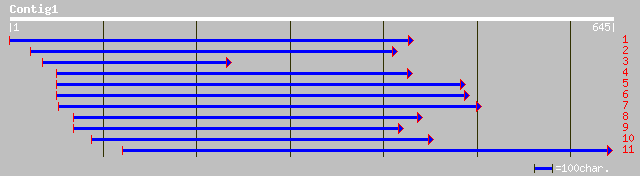
emb|CAA07229.2| putative beta-amilase [Cicer arietinum] 136 3e-31
gb|AAD38148.1|AF139501_1 beta-amylase [Prunus armeniaca] 102 3e-21
gb|AAK85300.1|AF402598_1 putative beta-amylase BMY3 [Arabidopsis... 74 1e-12
ref|NP_197368.1| glycosyl hydrolase family 14 (beta-amylase); pr... 74 1e-12
dbj|BAC20078.1| putative beta-amylase [Oryza sativa (japonica cu... 42 0.007
>emb|CAA07229.2| putative beta-amilase [Cicer arietinum]
Length = 314
Score = 136 bits (342), Expect = 3e-31
Identities = 68/86 (79%), Positives = 75/86 (87%), Gaps = 2/86 (2%)
Frame = -1
Query: 616 SVFGAPKGFDQIKKNLSGDNVLDLFPYQRMGAYFFFPEHFPSFTEFVRSLNQPELHSDDL 437
S FG+P GF+QIKKN+SGDNVLDLF YQRMGAYFF PEHFPSFTE VRS+NQP+LH DDL
Sbjct: 229 SEFGSPGGFEQIKKNISGDNVLDLFTYQRMGAYFFSPEHFPSFTELVRSVNQPKLHFDDL 288
Query: 436 PTVEEE--GTESAVNSQESSISMQAA 365
PT EEE G E+AV SQESS+SMQAA
Sbjct: 289 PTEEEEGGGGETAVMSQESSVSMQAA 314
>gb|AAD38148.1|AF139501_1 beta-amylase [Prunus armeniaca]
Length = 450
Score = 102 bits (255), Expect = 3e-21
Identities = 54/84 (64%), Positives = 62/84 (73%)
Frame = -1
Query: 616 SVFGAPKGFDQIKKNLSGDNVLDLFPYQRMGAYFFFPEHFPSFTEFVRSLNQPELHSDDL 437
SV G GF QIKKNL G+NV+DLF YQRMGA FF PEHFP F++FV +LNQP L SDDL
Sbjct: 368 SVSGGHGGFQQIKKNLMGENVMDLFTYQRMGADFFSPEHFPLFSKFVWTLNQPALQSDDL 427
Query: 436 PTVEEEGTESAVNSQESSISMQAA 365
P +EEE ES ++ ES MQAA
Sbjct: 428 P-IEEEVVESVRSNSESVTHMQAA 450
>gb|AAK85300.1|AF402598_1 putative beta-amylase BMY3 [Arabidopsis thaliana]
Length = 537
Score = 74.3 bits (181), Expect = 1e-12
Identities = 41/81 (50%), Positives = 55/81 (67%), Gaps = 2/81 (2%)
Frame = -1
Query: 601 PKGFDQIKKNLSGDNV-LDLFPYQRMGAYFFFPEHFPSFTEFVRSLNQPELHSDDLPT-V 428
P GF++I +NL +NV +DLF YQRMGA FF PEHF +FT FVR+L+Q EL SDD +
Sbjct: 457 PGGFERIVENLKDENVGIDLFTYQRMGALFFSPEHFHAFTVFVRNLSQFELSSDDQASEA 516
Query: 427 EEEGTESAVNSQESSISMQAA 365
E E +++ S + S+Q A
Sbjct: 517 EVEAETASIGSGTGAPSLQTA 537
>ref|NP_197368.1| glycosyl hydrolase family 14 (beta-amylase); protein id:
At5g18670.1, supported by cDNA: 30798., supported by
cDNA: gi_15149456, supported by cDNA: gi_17978934
[Arabidopsis thaliana] gi|17978935|gb|AAL47434.1|
AT5g18670/T1A4_50 [Arabidopsis thaliana]
gi|21592648|gb|AAM64597.1| beta-amylase-like proten
[Arabidopsis thaliana] gi|22655358|gb|AAM98271.1|
At5g18670/T1A4_50 [Arabidopsis thaliana]
Length = 536
Score = 74.3 bits (181), Expect = 1e-12
Identities = 41/81 (50%), Positives = 55/81 (67%), Gaps = 2/81 (2%)
Frame = -1
Query: 601 PKGFDQIKKNLSGDNV-LDLFPYQRMGAYFFFPEHFPSFTEFVRSLNQPELHSDDLPT-V 428
P GF++I +NL +NV +DLF YQRMGA FF PEHF +FT FVR+L+Q EL SDD +
Sbjct: 456 PGGFERIVENLKDENVGIDLFTYQRMGALFFSPEHFHAFTVFVRNLSQFELSSDDQASEA 515
Query: 427 EEEGTESAVNSQESSISMQAA 365
E E +++ S + S+Q A
Sbjct: 516 EVEAETASIGSGTGAPSLQTA 536
>dbj|BAC20078.1| putative beta-amylase [Oryza sativa (japonica cultivar-group)]
Length = 523
Score = 42.0 bits (97), Expect = 0.007
Identities = 19/35 (54%), Positives = 23/35 (65%)
Frame = -1
Query: 544 FPYQRMGAYFFFPEHFPSFTEFVRSLNQPELHSDD 440
F YQRMG FF PEH+P+F EFVR + E +D
Sbjct: 472 FTYQRMGEAFFSPEHWPAFVEFVRGVVCGEWPDED 506
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 524,371,692
Number of Sequences: 1393205
Number of extensions: 10892941
Number of successful extensions: 24928
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 24031
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24885
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)