Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001592A_C02 KMC001592A_c02
(662 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
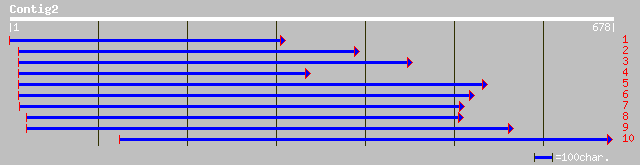
dbj|BAB62405.1| mitochondrial processing peptidase alpha subunit... 143 2e-33
sp|P29677|MPPA_SOLTU Mitochondrial processing peptidase alpha su... 139 3e-32
pir||S51590 mitochondrial processing peptidase (EC 3.4.24.64) al... 135 5e-31
ref|NP_175610.1| mitochondrial processing peptidase alpha subuni... 131 7e-30
gb|AAK59675.1| putative mitochondrial processing peptidase alpha... 130 1e-29
>dbj|BAB62405.1| mitochondrial processing peptidase alpha subunit [Morus alba]
Length = 506
Score = 143 bits (360), Expect = 2e-33
Identities = 69/108 (63%), Positives = 88/108 (80%)
Frame = -3
Query: 645 SDFVSKVIDITVNELLAVATSGPVNHVELDRAQPATQSAILMNLESRMVVSEDIGRQVLT 466
SDF +K +D+ NEL+A++ G V+ V+LDRA+ +T+SAILMNLESR++ SEDIGRQVLT
Sbjct: 389 SDFAAKAVDVVANELIAISKPGEVDQVQLDRAKKSTKSAILMNLESRVIASEDIGRQVLT 448
Query: 465 YAERKPVDDFLKAVDQVTLKDITSISHKLLSSPLPMASYGDVLYVPSY 322
Y +R VD FL AVD+VT+KDI S + KLLSSP+ +ASYGDVLY PSY
Sbjct: 449 YGKRMEVDHFLNAVDEVTVKDIASTAQKLLSSPVTLASYGDVLYFPSY 496
>sp|P29677|MPPA_SOLTU Mitochondrial processing peptidase alpha subunit, mitochondrial
precursor (Alpha-MPP) (Ubiquinol-cytochrome C reductase
subunit II) gi|421956|pir||S23558 ubiquinol-cytochrome-c
reductase (EC 1.10.2.2) alpha chain precursor - potato
gi|21493|emb|CAA46990.1| mitochondrial processing
peptidase [Solanum tuberosum]
Length = 504
Score = 139 bits (350), Expect = 3e-32
Identities = 70/108 (64%), Positives = 86/108 (78%)
Frame = -3
Query: 645 SDFVSKVIDITVNELLAVATSGPVNHVELDRAQPATQSAILMNLESRMVVSEDIGRQVLT 466
SDF + +D+ V EL+AVA V+ V+L+RA+ AT+SAILMNLESRMV SEDIGRQ+LT
Sbjct: 387 SDFGPQAVDVAVKELIAVANPSEVDQVQLNRAKQATKSAILMNLESRMVASEDIGRQLLT 446
Query: 465 YAERKPVDDFLKAVDQVTLKDITSISHKLLSSPLPMASYGDVLYVPSY 322
Y ER PV+ FLKA+D V+ KDI S+ KL+SSPL MASYGDVL +PSY
Sbjct: 447 YGERNPVEHFLKAIDAVSAKDIASVVQKLISSPLTMASYGDVLSLPSY 494
>pir||S51590 mitochondrial processing peptidase (EC 3.4.24.64) alpha-II chain
precursor - potato gi|587562|emb|CAA56520.1|
mitochondrial processing peptidase [Solanum tuberosum]
Length = 504
Score = 135 bits (340), Expect = 5e-31
Identities = 67/108 (62%), Positives = 85/108 (78%)
Frame = -3
Query: 645 SDFVSKVIDITVNELLAVATSGPVNHVELDRAQPATQSAILMNLESRMVVSEDIGRQVLT 466
SDF + I++ V EL AVA G V+ V+LDRA+ +T+SAILMNLESRMV SEDIGRQ+L
Sbjct: 387 SDFAPRAIEVAVKELTAVANPGEVDMVQLDRAKQSTKSAILMNLESRMVASEDIGRQLLI 446
Query: 465 YAERKPVDDFLKAVDQVTLKDITSISHKLLSSPLPMASYGDVLYVPSY 322
Y ERKPV+ LKA+D ++ DI S++ KL+SSPL MASYGDVL +P+Y
Sbjct: 447 YGERKPVEHVLKAIDAISANDIASVAQKLISSPLTMASYGDVLSLPTY 494
>ref|NP_175610.1| mitochondrial processing peptidase alpha subunit, putative; protein
id: At1g51980.1, supported by cDNA: 6390., supported by
cDNA: gi_17529269, supported by cDNA: gi_20258956
[Arabidopsis thaliana] gi|25290029|pir||D96559
hypothetical protein F5F19.4 [imported] - Arabidopsis
thaliana gi|4220446|gb|AAD12673.1| Strong similarity to
gi|2062155 T02O04.2 mitochondrial processing peptidase
alpha subunit precusor isolog from Arabidopsis thaliana
BAC gb|AC001645. ESTs gb|Z18504 and gb|AA395715 come
from this gene gi|17529270|gb|AAL38862.1| putative
mitochondrial processing peptidase alpha subunit
[Arabidopsis thaliana] gi|20258957|gb|AAM14194.1|
putative mitochondrial processing peptidase alpha
subunit [Arabidopsis thaliana]
Length = 503
Score = 131 bits (330), Expect = 7e-30
Identities = 66/106 (62%), Positives = 81/106 (76%)
Frame = -3
Query: 639 FVSKVIDITVNELLAVATSGPVNHVELDRAQPATQSAILMNLESRMVVSEDIGRQVLTYA 460
F +K I++ EL VA G VN LDRA+ AT+SA+LMNLESRM+ +EDIGRQ+LTY
Sbjct: 391 FAAKAIELAAKELKDVA-GGKVNQAHLDRAKAATKSAVLMNLESRMIAAEDIGRQILTYG 449
Query: 459 ERKPVDDFLKAVDQVTLKDITSISHKLLSSPLPMASYGDVLYVPSY 322
ERKPVD FLK+VDQ+TLKDI + K++S PL M S+GDVL VPSY
Sbjct: 450 ERKPVDQFLKSVDQLTLKDIADFTSKVISKPLTMGSFGDVLAVPSY 495
>gb|AAK59675.1| putative mitochondrial processing peptidase alpha subunit
[Arabidopsis thaliana]
Length = 499
Score = 130 bits (328), Expect = 1e-29
Identities = 66/107 (61%), Positives = 84/107 (77%)
Frame = -3
Query: 642 DFVSKVIDITVNELLAVATSGPVNHVELDRAQPATQSAILMNLESRMVVSEDIGRQVLTY 463
+F S+ I++ +E+ AVA G VN LDRA+ AT+SAILMNLESRM+ +EDIGRQ+LTY
Sbjct: 386 EFASQGIELVASEMNAVA-DGKVNQKHLDRAKAATKSAILMNLESRMIAAEDIGRQILTY 444
Query: 462 AERKPVDDFLKAVDQVTLKDITSISHKLLSSPLPMASYGDVLYVPSY 322
ERKPVD FLK VDQ+TLKDI + K+++ PL MA++GDVL VPSY
Sbjct: 445 GERKPVDQFLKTVDQLTLKDIADFTSKVITKPLTMATFGDVLNVPSY 491
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 575,512,742
Number of Sequences: 1393205
Number of extensions: 12456804
Number of successful extensions: 25938
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 25340
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25933
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28572683052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)