Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001591A_C03 KMC001591A_c03
(582 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
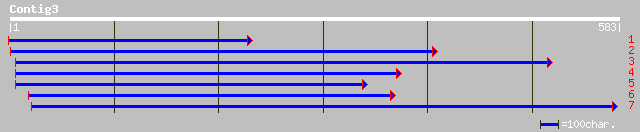
ref|NP_190855.1| putative protein; protein id: At3g52870.1 [Arab... 112 3e-24
gb|AAM78112.1| AT3g52870/F8J2_40 [Arabidopsis thaliana] 112 3e-24
ref|NP_187969.1| hypothetical protein; protein id: At3g13600.1 [... 91 1e-17
dbj|BAC07110.1| hypothetical protein~similar to Arabidopsis thal... 86 4e-16
ref|NP_191407.1| putative protein; protein id: At3g58480.1 [Arab... 74 1e-12
>ref|NP_190855.1| putative protein; protein id: At3g52870.1 [Arabidopsis thaliana]
gi|11292181|pir||T47544 hypothetical protein F8J2.40 -
Arabidopsis thaliana gi|7529711|emb|CAB86891.1| putative
protein [Arabidopsis thaliana]
gi|24111367|gb|AAN46807.1| At3g52870/F8J2_40
[Arabidopsis thaliana]
Length = 456
Score = 112 bits (281), Expect = 3e-24
Identities = 56/81 (69%), Positives = 66/81 (81%)
Frame = -2
Query: 566 SYKRTLSGGLQSPRAAEVPKTAILQRINSKKATKSYQLGHNLSRKWSTGAGPRIGCVADY 387
SY+RTLSGGL SP+A VP+ ++L RINSKK ++S QLGH LS KWSTG GPRIGC ADY
Sbjct: 367 SYQRTLSGGLGSPKA-NVPQKSMLLRINSKKQSRSLQLGHQLSLKWSTGVGPRIGCAADY 425
Query: 386 PVELRWQALEMLNLSPKFPPS 324
PV+LR QALE +NLSPK+ S
Sbjct: 426 PVQLRTQALEFVNLSPKYRSS 446
>gb|AAM78112.1| AT3g52870/F8J2_40 [Arabidopsis thaliana]
Length = 456
Score = 112 bits (281), Expect = 3e-24
Identities = 56/81 (69%), Positives = 66/81 (81%)
Frame = -2
Query: 566 SYKRTLSGGLQSPRAAEVPKTAILQRINSKKATKSYQLGHNLSRKWSTGAGPRIGCVADY 387
SY+RTLSGGL SP+A VP+ ++L RINSKK ++S QLGH LS KWSTG GPRIGC ADY
Sbjct: 367 SYQRTLSGGLGSPKA-NVPQKSMLLRINSKKQSRSLQLGHQLSLKWSTGVGPRIGCAADY 425
Query: 386 PVELRWQALEMLNLSPKFPPS 324
PV+LR QALE +NLSPK+ S
Sbjct: 426 PVQLRTQALEFVNLSPKYRSS 446
>ref|NP_187969.1| hypothetical protein; protein id: At3g13600.1 [Arabidopsis
thaliana] gi|11994562|dbj|BAB02602.1|
emb|CAB68186.1~gene_id:K20M4.4~strong similarity to
unknown protein [Arabidopsis thaliana]
Length = 605
Score = 90.5 bits (223), Expect = 1e-17
Identities = 42/70 (60%), Positives = 54/70 (77%)
Frame = -2
Query: 545 GGLQSPRAAEVPKTAILQRINSKKATKSYQLGHNLSRKWSTGAGPRIGCVADYPVELRWQ 366
G + ++ + +IL+RINSKK TKS+QLG LS KW+TGAGPRIGCV DYP EL++Q
Sbjct: 498 GETKESEVVKITEESILKRINSKKETKSFQLGKQLSCKWTTGAGPRIGCVRDYPSELQFQ 557
Query: 365 ALEMLNLSPK 336
ALE +NLSP+
Sbjct: 558 ALEQVNLSPR 567
>dbj|BAC07110.1| hypothetical protein~similar to Arabidopsis thaliana chromosome 3,
At3g13600 [Oryza sativa (japonica cultivar-group)]
gi|22296385|dbj|BAC10154.1| P0519E12.30 [Oryza sativa
(japonica cultivar-group)]
Length = 585
Score = 85.5 bits (210), Expect = 4e-16
Identities = 43/86 (50%), Positives = 56/86 (65%)
Frame = -2
Query: 524 AAEVPKTAILQRINSKKATKSYQLGHNLSRKWSTGAGPRIGCVADYPVELRWQALEMLNL 345
++ VP+ IL+RINSKK KSYQLG LS KW+TGAGPRI CV DYP EL+ +ALE ++L
Sbjct: 474 SSSVPREKILERINSKKEAKSYQLGKQLSFKWTTGAGPRIVCVRDYPSELQLRALEQVHL 533
Query: 344 SPKFPPSPSSYMLMGGLVSPAACPTP 267
SP+ + + SP +P
Sbjct: 534 SPRSAAAAAGGRPSSRFASPQRSSSP 559
>ref|NP_191407.1| putative protein; protein id: At3g58480.1 [Arabidopsis thaliana]
gi|11357446|pir||T45668 hypothetical protein F14P22.70 -
Arabidopsis thaliana gi|6735365|emb|CAB68186.1| putative
protein [Arabidopsis thaliana]
Length = 575
Score = 74.3 bits (181), Expect = 1e-12
Identities = 40/86 (46%), Positives = 52/86 (59%)
Frame = -2
Query: 515 VPKTAILQRINSKKATKSYQLGHNLSRKWSTGAGPRIGCVADYPVELRWQALEMLNLSPK 336
V K I++RI+S K KSYQL L +WSTGAGPRI C+ DYP EL+++ LE +LSP+
Sbjct: 483 VLKEKIMRRIDSHKGIKSYQLAERLHSRWSTGAGPRISCMRDYPSELQFRVLEQAHLSPR 542
Query: 335 FPPSPSSYMLMGGLVSPAACPTPRGT 258
+ S +SP A RGT
Sbjct: 543 ASSNSSK-------ISPFAPVRTRGT 561
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 527,184,477
Number of Sequences: 1393205
Number of extensions: 11871077
Number of successful extensions: 30175
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 28913
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30161
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)