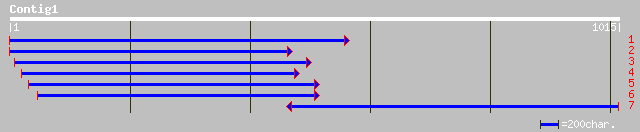
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001570A_C01 KMC001570A_c01
(1015 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P52427|PSA4_SPIOL Proteasome subunit alpha type 4 (20S protea... 457 e-128
ref|NP_188850.1| 20S proteasome alpha subunit C (PAC1); protein ... 455 e-127
emb|CAA73624.1| multicatalytic endopeptidase [Arabidopsis thaliana] 453 e-126
sp|O82530|PSA4_PETHY Proteasome subunit alpha type 4 (20S protea... 448 e-125
gb|AAF34770.1|AF227625_1 proteasome 27 kDa subunit [Euphorbia es... 441 e-122
>sp|P52427|PSA4_SPIOL Proteasome subunit alpha type 4 (20S proteasome alpha subunit C)
(20S proteasome subunit alpha-3) (Proteasome 27 kDa
subunit) gi|7435866|pir||T09160 proteasome subunit -
spinach gi|1262146|emb|CAA65660.1| proteasome subunit
[Spinacia oleracea]
Length = 250
Score = 457 bits (1177), Expect = e-128
Identities = 227/250 (90%), Positives = 243/250 (96%)
Frame = -2
Query: 948 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGAAIGILSKDGVVLVGEKKVTSKLLQTST 769
MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAG+AIGIL+KDGVVL+GEKKVTSKLLQTST
Sbjct: 1 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGSAIGILAKDGVVLIGEKKVTSKLLQTST 60
Query: 768 STEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFAYQEPMPVEQLVQSLCDTKQG 589
STEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTF+YQEPMPVEQLVQSLCDTKQG
Sbjct: 61 STEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFSYQEPMPVEQLVQSLCDTKQG 120
Query: 588 YTQFGGLRPFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKAGAIGANNQAAQSILKQDYK 409
YTQFGGLRPFGVSFLFAGWDKN+GFQLYMSDPSGNYGGWKA AIGANNQAAQS+LKQDYK
Sbjct: 121 YTQFGGLRPFGVSFLFAGWDKNYGFQLYMSDPSGNYGGWKATAIGANNQAAQSMLKQDYK 180
Query: 408 DDITREEAVQLALKVLSKTMDSTSLTSDKLELAEVYLSPSGKVKYEVCSPESLTKLLVKS 229
DD+TRE+AV+LALK LSKTMDSTSLTS+KLELAEVYL PSGKVKY+V SPESL +LL +S
Sbjct: 181 DDVTREDAVKLALKALSKTMDSTSLTSEKLELAEVYLLPSGKVKYQVHSPESLNRLLTES 240
Query: 228 GVTQPATESA 199
G+TQPA E++
Sbjct: 241 GLTQPAAETS 250
>ref|NP_188850.1| 20S proteasome alpha subunit C (PAC1); protein id: At3g22110.1,
supported by cDNA: 19620., supported by cDNA:
gi_14423431, supported by cDNA: gi_3421076 [Arabidopsis
thaliana] gi|12643237|sp|O81148|PSA4_ARATH Proteasome
subunit alpha type 4 (20S proteasome alpha subunit C)
(Proteasome 27 kDa subunit) gi|25290058|pir||T51969 20S
proteasome subunit PAC1 [imported] - Arabidopsis
thaliana gi|3421077|gb|AAC32057.1| 20S proteasome
subunit PAC1 [Arabidopsis thaliana]
gi|11994731|dbj|BAB03060.1| 20S proteasome subunit PAC1
[Arabidopsis thaliana]
gi|14423432|gb|AAK62398.1|AF386953_1 20S proteasome
subunit PAC1 [Arabidopsis thaliana]
gi|21554045|gb|AAM63126.1| 20S proteasome subunit PAC1
[Arabidopsis thaliana] gi|23197586|gb|AAN15320.1| 20S
proteasome subunit PAC1 [Arabidopsis thaliana]
Length = 250
Score = 455 bits (1170), Expect = e-127
Identities = 229/250 (91%), Positives = 239/250 (95%)
Frame = -2
Query: 948 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGAAIGILSKDGVVLVGEKKVTSKLLQTST 769
MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAG+AIGILSKDGVVL+GEKKVTSKLLQTST
Sbjct: 1 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGSAIGILSKDGVVLIGEKKVTSKLLQTST 60
Query: 768 STEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFAYQEPMPVEQLVQSLCDTKQG 589
S EKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTF YQEPMPVEQLVQSLCDTKQG
Sbjct: 61 SAEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFMYQEPMPVEQLVQSLCDTKQG 120
Query: 588 YTQFGGLRPFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKAGAIGANNQAAQSILKQDYK 409
YTQFGGLRPFGVSFLFAGWDK+ GFQLYMSDPSGNYGGWKA A+GANNQAAQSILKQDYK
Sbjct: 121 YTQFGGLRPFGVSFLFAGWDKHHGFQLYMSDPSGNYGGWKAAAVGANNQAAQSILKQDYK 180
Query: 408 DDITREEAVQLALKVLSKTMDSTSLTSDKLELAEVYLSPSGKVKYEVCSPESLTKLLVKS 229
DD TREEAV+LALKVL+KTMDSTSLTS+KLELAEVYL+PS VKY V SPESLTKLLVK
Sbjct: 181 DDATREEAVELALKVLTKTMDSTSLTSEKLELAEVYLTPSKTVKYHVHSPESLTKLLVKH 240
Query: 228 GVTQPATESA 199
GVTQPA E++
Sbjct: 241 GVTQPAAETS 250
>emb|CAA73624.1| multicatalytic endopeptidase [Arabidopsis thaliana]
Length = 250
Score = 453 bits (1166), Expect = e-126
Identities = 228/250 (91%), Positives = 238/250 (95%)
Frame = -2
Query: 948 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGAAIGILSKDGVVLVGEKKVTSKLLQTST 769
MSRRYDSRTTIFSPEGRLYQVEY MEAIGNAG+AIGILSKDGVVL+GEKKVTSKLLQTST
Sbjct: 1 MSRRYDSRTTIFSPEGRLYQVEYVMEAIGNAGSAIGILSKDGVVLIGEKKVTSKLLQTST 60
Query: 768 STEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFAYQEPMPVEQLVQSLCDTKQG 589
S EKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTF YQEPMPVEQLVQSLCDTKQG
Sbjct: 61 SAEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFMYQEPMPVEQLVQSLCDTKQG 120
Query: 588 YTQFGGLRPFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKAGAIGANNQAAQSILKQDYK 409
YTQFGGLRPFGVSFLFAGWDK+ GFQLYMSDPSGNYGGWKA A+GANNQAAQSILKQDYK
Sbjct: 121 YTQFGGLRPFGVSFLFAGWDKHHGFQLYMSDPSGNYGGWKAAAVGANNQAAQSILKQDYK 180
Query: 408 DDITREEAVQLALKVLSKTMDSTSLTSDKLELAEVYLSPSGKVKYEVCSPESLTKLLVKS 229
DD TREEAV+LALKVL+KTMDSTSLTS+KLELAEVYL+PS VKY V SPESLTKLLVK
Sbjct: 181 DDATREEAVELALKVLTKTMDSTSLTSEKLELAEVYLTPSKTVKYHVHSPESLTKLLVKH 240
Query: 228 GVTQPATESA 199
GVTQPA E++
Sbjct: 241 GVTQPAAETS 250
>sp|O82530|PSA4_PETHY Proteasome subunit alpha type 4 (20S proteasome alpha subunit C)
(20S proteasome subunit alpha-3)
gi|3608483|gb|AAC35982.1| proteasome alpha subunit
[Petunia x hybrida]
Length = 249
Score = 448 bits (1152), Expect = e-125
Identities = 225/250 (90%), Positives = 239/250 (95%)
Frame = -2
Query: 948 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGAAIGILSKDGVVLVGEKKVTSKLLQTST 769
MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAG+AIGI SKDGVVLVGEKKVTSKLLQTST
Sbjct: 1 MSRRYDSRTTIFSPEGRLYQVEYAMEAIGNAGSAIGISSKDGVVLVGEKKVTSKLLQTST 60
Query: 768 STEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFAYQEPMPVEQLVQSLCDTKQG 589
S+EKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTF+YQEPMPVEQLVQSLCDTKQG
Sbjct: 61 SSEKMYKIDDHVACAVAGIMSDANILINTARVQAQRYTFSYQEPMPVEQLVQSLCDTKQG 120
Query: 588 YTQFGGLRPFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKAGAIGANNQAAQSILKQDYK 409
YTQ+GGL PFGVSFLFAGWDKNFGFQL+MSDPSGNY GWKA AIGANNQAAQS+LKQDYK
Sbjct: 121 YTQYGGLPPFGVSFLFAGWDKNFGFQLFMSDPSGNYAGWKAAAIGANNQAAQSMLKQDYK 180
Query: 408 DDITREEAVQLALKVLSKTMDSTSLTSDKLELAEVYLSPSGKVKYEVCSPESLTKLLVKS 229
DDITREEAVQLALKVLSKTMDSTSLTS+KLELAEV+LS +GKVKY+ CSPE L +LVKS
Sbjct: 181 DDITREEAVQLALKVLSKTMDSTSLTSEKLELAEVFLS-NGKVKYQACSPEKLNSMLVKS 239
Query: 228 GVTQPATESA 199
G+TQP+ E +
Sbjct: 240 GLTQPSAEES 249
>gb|AAF34770.1|AF227625_1 proteasome 27 kDa subunit [Euphorbia esula]
Length = 242
Score = 441 bits (1133), Expect = e-122
Identities = 219/242 (90%), Positives = 234/242 (96%)
Frame = -2
Query: 924 TTIFSPEGRLYQVEYAMEAIGNAGAAIGILSKDGVVLVGEKKVTSKLLQTSTSTEKMYKI 745
TTIFSPEGRLYQVEYAMEAIGNAG AIGILSKDGVVLVGEKKVTSKLLQTSTSTEKMYKI
Sbjct: 1 TTIFSPEGRLYQVEYAMEAIGNAGTAIGILSKDGVVLVGEKKVTSKLLQTSTSTEKMYKI 60
Query: 744 DDHVACAVAGIMSDANILINTARVQAQRYTFAYQEPMPVEQLVQSLCDTKQGYTQFGGLR 565
DDHV+CAVAGIMSDANILINTARVQAQRY F+YQEPMPVEQLVQSLCDTKQGYTQFGGLR
Sbjct: 61 DDHVSCAVAGIMSDANILINTARVQAQRYYFSYQEPMPVEQLVQSLCDTKQGYTQFGGLR 120
Query: 564 PFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKAGAIGANNQAAQSILKQDYKDDITREEA 385
PFGVSF+FAGWDKNFGFQLY SDPSGNYGGWKA AIGANNQAAQ +LKQDYKD+ITREEA
Sbjct: 121 PFGVSFIFAGWDKNFGFQLYTSDPSGNYGGWKAAAIGANNQAAQFMLKQDYKDEITREEA 180
Query: 384 VQLALKVLSKTMDSTSLTSDKLELAEVYLSPSGKVKYEVCSPESLTKLLVKSGVTQPATE 205
VQLALKVLSKTMDSTSLTSDKLELAEV+L PSGKVKY+VCSPE+++KLLV+ GVTQP+ +
Sbjct: 181 VQLALKVLSKTMDSTSLTSDKLELAEVFLLPSGKVKYQVCSPETVSKLLVQYGVTQPSAD 240
Query: 204 SA 199
++
Sbjct: 241 AS 242
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 918,380,306
Number of Sequences: 1393205
Number of extensions: 21472664
Number of successful extensions: 55506
Number of sequences better than 10.0: 544
Number of HSP's better than 10.0 without gapping: 51903
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 54933
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 58773479151
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)