Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001567A_C05 KMC001567A_c05
(730 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
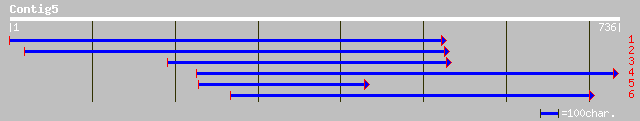
ref|XP_233644.1| tumor necrosis factor receptor superfamily, mem... 33 4.1
sp|Q33446|GATB_EMENI Probable glutamyl-tRNA(Gln) amidotransferas... 33 4.1
ref|XP_236375.1| similar to RIKEN cDNA 2010013H22 gene [Mus musc... 33 5.4
ref|NP_349368.1| Membrane assosiated methyl-accepting chemotaxis... 33 5.4
ref|NP_775434.1| beta-1,6-N-acetylglucosaminyltransferase [Rattu... 33 5.4
>ref|XP_233644.1| tumor necrosis factor receptor superfamily, member 1b [Rattus
norvegicus]
Length = 521
Score = 33.1 bits (74), Expect = 4.1
Identities = 21/61 (34%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Frame = -1
Query: 658 VTQKKQNAGCAQKKSMVEHTQEDHEAHSWEHQIQYPVKSLDNV---QLHVFSEAPSSSSS 488
+ Q+K+ C Q+++MV H +D KS D + Q H+ + APSSSSS
Sbjct: 332 LVQRKKKPSCLQRETMVPHLPDD--------------KSQDAIGLEQQHLLTTAPSSSSS 377
Query: 487 S 485
S
Sbjct: 378 S 378
>sp|Q33446|GATB_EMENI Probable glutamyl-tRNA(Gln) amidotransferase subunit B,
mitochondrial precursor (GLU-ADT subunit B) (NEMPA
protein) gi|1408310|gb|AAB05037.1| NEMPA
Length = 581
Score = 33.1 bits (74), Expect = 4.1
Identities = 16/54 (29%), Positives = 25/54 (45%)
Frame = -1
Query: 652 QKKQNAGCAQKKSMVEHTQEDHEAHSWEHQIQYPVKSLDNVQLHVFSEAPSSSS 491
Q KQ A + + QE+ H WE + + + N + +FS AP+S S
Sbjct: 65 QLKQEAKAIKSRKRERREQEEASRHKWELTVGVEIHAQLNTETKLFSRAPTSPS 118
>ref|XP_236375.1| similar to RIKEN cDNA 2010013H22 gene [Mus musculus] [Rattus
norvegicus]
Length = 626
Score = 32.7 bits (73), Expect = 5.4
Identities = 13/38 (34%), Positives = 24/38 (62%)
Frame = +3
Query: 444 NLRNLARIGLWQEDEDELEDGASLNTCSCTLSRLLTGY 557
++R++AR+ WQ+ E ++E+GA +CS R + Y
Sbjct: 544 DMRSIARLTKWQDHEGDIENGAPYTSCSGIHQRAICVY 581
>ref|NP_349368.1| Membrane assosiated methyl-accepting chemotaxis protein with HAMP
domain [Clostridium acetobutylicum]
gi|25496115|pir||A97240 membrane assosiated
methyl-accepting chemotaxis protein with HAMP domain
[imported] - Clostridium acetobutylicum
gi|15025801|gb|AAK80708.1|AE007774_5 Membrane assosiated
methyl-accepting chemotaxis protein with HAMP domain
[Clostridium acetobutylicum]
Length = 571
Score = 32.7 bits (73), Expect = 5.4
Identities = 17/54 (31%), Positives = 31/54 (56%), Gaps = 2/54 (3%)
Frame = -1
Query: 286 IRIVIIYGGVV--LKIENKNKHLCHIWSISLLNTFHLYSFHGHMLCYRDDLTLV 131
+ ++ GVV LKI NK+L +I+++ + T LY G+++ R D+ L+
Sbjct: 20 VAVISTIAGVVGVLKINGVNKNLDNIYNVDMKGTNILYQIRGYVIEVRADILLI 73
>ref|NP_775434.1| beta-1,6-N-acetylglucosaminyltransferase [Rattus norvegicus]
gi|27372228|dbj|BAC53607.1|
beta-1,6-N-acetylglucosaminyltransferase [Rattus
norvegicus]
Length = 437
Score = 32.7 bits (73), Expect = 5.4
Identities = 13/38 (34%), Positives = 24/38 (62%)
Frame = +3
Query: 444 NLRNLARIGLWQEDEDELEDGASLNTCSCTLSRLLTGY 557
++R++AR+ WQ+ E ++E+GA +CS R + Y
Sbjct: 355 DMRSIARLTKWQDHEGDIENGAPYTSCSGIHQRAICVY 392
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 634,306,547
Number of Sequences: 1393205
Number of extensions: 13842726
Number of successful extensions: 35602
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 33378
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35440
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34343566934
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)