Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001554A_C01 KMC001554A_c01
(614 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
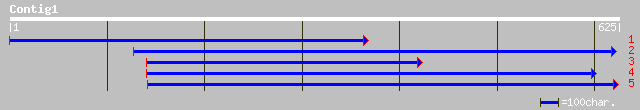
sp|O22307|C7DB_LOTJA Cytochrome P450 71D11 gi|2367431|gb|AAB6964... 283 2e-75
sp|O81971|C7D9_SOYBN Cytochrome P450 71D9 (P450 CP3) gi|7430686|... 190 1e-47
gb|AAO32823.1| cytochrome P450 71D2 [Catharanthus roseus] 171 9e-42
sp|O48923|C7DA_SOYBN Cytochrome P450 71D10 gi|7430682|pir||T0593... 170 2e-41
sp|P93531|C7D7_SOLCH Cytochrome P450 71D7 gi|7430645|pir||T10499... 164 7e-40
>sp|O22307|C7DB_LOTJA Cytochrome P450 71D11 gi|2367431|gb|AAB69644.1| putative cytochrome
P450 [Lotus japonicus]
Length = 490
Score = 283 bits (723), Expect = 2e-75
Identities = 133/134 (99%), Positives = 134/134 (99%)
Frame = -2
Query: 613 VFRECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDYQGTNFE 434
VFRECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDY+GTNFE
Sbjct: 357 VFRECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDYKGTNFE 416
Query: 433 HLPFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKED 254
HLPFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKED
Sbjct: 417 HLPFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKED 476
Query: 253 LCLIPSISHPLPST 212
LCLIPSISHPLPST
Sbjct: 477 LCLIPSISHPLPST 490
>sp|O81971|C7D9_SOYBN Cytochrome P450 71D9 (P450 CP3) gi|7430686|pir||T07117 probable
cytochrome P450 CP3 - soybean gi|3334661|emb|CAA71514.1|
putative cytochrome P450 [Glycine max]
Length = 496
Score = 190 bits (482), Expect = 1e-47
Identities = 84/127 (66%), Positives = 104/127 (81%)
Frame = -2
Query: 607 RECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDYQGTNFEHL 428
REC QACEINGYHIPAKS V+VN +AIG D + W E ERF PERFI+ SI+Y+ +FE +
Sbjct: 369 RECGQACEINGYHIPAKSRVIVNAWAIGRDPRLWTEAERFYPERFIERSIEYKSNSFEFI 428
Query: 427 PFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKEDLC 248
PFGAGRR+CPG+ +G++NVE VLA+L+YHFDW LPKG KNEDL +TE FG+TV++K+DL
Sbjct: 429 PFGAGRRMCPGLTFGLSNVEYVLAMLMYHFDWKLPKGTKNEDLGMTEIFGITVARKDDLY 488
Query: 247 LIPSISH 227
LIP H
Sbjct: 489 LIPKTVH 495
>gb|AAO32823.1| cytochrome P450 71D2 [Catharanthus roseus]
Length = 430
Score = 171 bits (432), Expect = 9e-42
Identities = 77/128 (60%), Positives = 96/128 (74%)
Frame = -2
Query: 607 RECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDYQGTNFEHL 428
RECR+AC+I+GY IP K+ V VN +AIG D KYW +PE F PERF D+S+D+ G NFE+L
Sbjct: 303 RECREACQIDGYDIPVKTRVFVNVWAIGRDEKYWKDPESFIPERFEDNSLDFTGNNFEYL 362
Query: 427 PFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKEDLC 248
PFG GRRICPG+ +G+ANV LVLALLLYHF+W LP G+ D+D+ E G+ SKK L
Sbjct: 363 PFGCGRRICPGMTFGLANVHLVLALLLYHFNWKLPPGV--NDIDMAERPGLGASKKHGLV 420
Query: 247 LIPSISHP 224
L+PS P
Sbjct: 421 LVPSFYQP 428
>sp|O48923|C7DA_SOYBN Cytochrome P450 71D10 gi|7430682|pir||T05939 cytochrome P450
monooxygenase 71D10p - soybean gi|2739000|gb|AAB94588.1|
CYP71D10p [Glycine max]
Length = 510
Score = 170 bits (430), Expect = 2e-41
Identities = 73/123 (59%), Positives = 98/123 (79%)
Frame = -2
Query: 607 RECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDYQGTNFEHL 428
R R+ C+INGY IP+K+ +++N +AIG + KYW E E F PERF++SSID++GT+FE +
Sbjct: 383 RVSRERCQINGYEIPSKTRIIINAWAIGRNPKYWGETESFKPERFLNSSIDFRGTDFEFI 442
Query: 427 PFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKEDLC 248
PFGAGRRICPGI + + N+EL LA LLYHFDW LP +KNE+LD+TE G+T+ ++ DLC
Sbjct: 443 PFGAGRRICPGITFAIPNIELPLAQLLYHFDWKLPNKMKNEELDMTESNGITLRRQNDLC 502
Query: 247 LIP 239
LIP
Sbjct: 503 LIP 505
>sp|P93531|C7D7_SOLCH Cytochrome P450 71D7 gi|7430645|pir||T10499 probable cytochrome
P450 (clone pGHgen) - Chaco potato
gi|1762144|gb|AAB61965.1| putative cytochrome P450
Length = 500
Score = 164 bits (416), Expect = 7e-40
Identities = 77/128 (60%), Positives = 95/128 (74%)
Frame = -2
Query: 607 RECRQACEINGYHIPAKSTVLVNTFAIGTDSKYWAEPERFCPERFIDSSIDYQGTNFEHL 428
RECR+ EINGY IP K+ V+VN +A+G D KYW + E F PERF SID+ G NFE+L
Sbjct: 373 RECREETEINGYTIPVKTKVMVNVWALGRDPKYWDDAESFKPERFEQCSIDFIGNNFEYL 432
Query: 427 PFGAGRRICPGINYGMANVELVLALLLYHFDWTLPKGIKNEDLDLTEEFGVTVSKKEDLC 248
PFG GRRICPGI++G+ANV L LA LLYHFDW LP G++ +DLDLTE G+T ++K DL
Sbjct: 433 PFGGGRRICPGISFGLANVYLPLAQLLYHFDWKLPTGMEPKDLDLTESAGITAARKGDLY 492
Query: 247 LIPSISHP 224
LI + P
Sbjct: 493 LIATPHQP 500
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 502,494,049
Number of Sequences: 1393205
Number of extensions: 10508147
Number of successful extensions: 33714
Number of sequences better than 10.0: 2624
Number of HSP's better than 10.0 without gapping: 31126
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32060
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24854530794
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)