Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001551A_C01 KMC001551A_c01
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
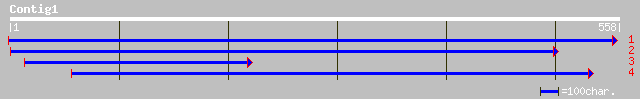
ref|NP_173104.1| unknown protein; protein id: At1g16560.1 [Arabi... 154 7e-37
dbj|BAB10175.1| gene_id:MTG10.16~unknown protein [Arabidopsis th... 113 1e-24
ref|NP_568951.1| putative protein; protein id: At5g62130.1, supp... 113 1e-24
gb|AAO00714.1| unknown protein [Oryza sativa (japonica cultivar-... 92 3e-18
gb|EAA35581.1| hypothetical protein [Neurospora crassa] 62 5e-09
>ref|NP_173104.1| unknown protein; protein id: At1g16560.1 [Arabidopsis thaliana]
gi|25372701|pir||H86300 F19K19.12 protein - Arabidopsis
thaliana gi|9989062|gb|AAG10825.1|AC011808_13 Unknown
protein [Arabidopsis thaliana]
Length = 342
Score = 154 bits (389), Expect = 7e-37
Identities = 68/90 (75%), Positives = 75/90 (82%)
Frame = -3
Query: 555 VCVVMAVVQLVIWAVWAGLSGHPSRWKLWLVVIAGGLAMLLEIYDFPPYEGLLDAHALWH 376
VCV M V QL +WA WA +S HPS WKLW+VVIAGGLAMLLEIYDFPPYEG DAH++WH
Sbjct: 249 VCVAMGVSQLFLWARWAAVSSHPSNWKLWVVVIAGGLAMLLEIYDFPPYEGYFDAHSIWH 308
Query: 375 ATTIPLTYIWWSFIRDDAEFRTSIRVKKAK 286
A TIPLT +WWSFIRDDAEFRTS +KK K
Sbjct: 309 AATIPLTILWWSFIRDDAEFRTSSLLKKTK 338
>dbj|BAB10175.1| gene_id:MTG10.16~unknown protein [Arabidopsis thaliana]
Length = 276
Score = 113 bits (283), Expect = 1e-24
Identities = 44/83 (53%), Positives = 64/83 (77%)
Frame = -3
Query: 534 VQLVIWAVWAGLSGHPSRWKLWLVVIAGGLAMLLEIYDFPPYEGLLDAHALWHATTIPLT 355
++LV+W +WA L+ HPS+WKL +I+ L + L ++DFPPY+G +DAHALW IPL+
Sbjct: 194 IELVVWGLWAALTSHPSKWKLRAFLISSILTLCLRMFDFPPYKGYIDAHALWRGAGIPLS 253
Query: 354 YIWWSFIRDDAEFRTSIRVKKAK 286
Y+WWSF+ DDA FRT++ +KK+K
Sbjct: 254 YLWWSFVCDDAVFRTTVNLKKSK 276
>ref|NP_568951.1| putative protein; protein id: At5g62130.1, supported by cDNA:
gi_15294203, supported by cDNA: gi_20147282 [Arabidopsis
thaliana] gi|15294204|gb|AAK95279.1|AF410293_1
AT5g62130/mtg10_150 [Arabidopsis thaliana]
gi|20147283|gb|AAM10355.1| AT5g62130/mtg10_150
[Arabidopsis thaliana]
Length = 343
Score = 113 bits (283), Expect = 1e-24
Identities = 44/83 (53%), Positives = 64/83 (77%)
Frame = -3
Query: 534 VQLVIWAVWAGLSGHPSRWKLWLVVIAGGLAMLLEIYDFPPYEGLLDAHALWHATTIPLT 355
++LV+W +WA L+ HPS+WKL +I+ L + L ++DFPPY+G +DAHALW IPL+
Sbjct: 261 IELVVWGLWAALTSHPSKWKLRAFLISSILTLCLRMFDFPPYKGYIDAHALWRGAGIPLS 320
Query: 354 YIWWSFIRDDAEFRTSIRVKKAK 286
Y+WWSF+ DDA FRT++ +KK+K
Sbjct: 321 YLWWSFVCDDAVFRTTVNLKKSK 343
>gb|AAO00714.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 347
Score = 92.4 bits (228), Expect = 3e-18
Identities = 39/90 (43%), Positives = 58/90 (64%)
Frame = -3
Query: 555 VCVVMAVVQLVIWAVWAGLSGHPSRWKLWLVVIAGGLAMLLEIYDFPPYEGLLDAHALWH 376
VC ++ Q ++WAVWA ++ HPS +K+ V+I +++LE YD PP G +D
Sbjct: 258 VCTAASLAQFLLWAVWAVMTKHPSCFKILFVIIGNVFSIVLETYDIPPRWGYVDGRVFCV 317
Query: 375 ATTIPLTYIWWSFIRDDAEFRTSIRVKKAK 286
A +IPLTY+WW F ++DAE RTS +KK +
Sbjct: 318 AISIPLTYLWWKFAKEDAEMRTSTIIKKTR 347
>gb|EAA35581.1| hypothetical protein [Neurospora crassa]
Length = 345
Score = 62.0 bits (149), Expect = 5e-09
Identities = 32/81 (39%), Positives = 46/81 (56%), Gaps = 4/81 (4%)
Frame = -3
Query: 549 VVMAVVQLVIWA--VWAGLSGHPSRWKLW--LVVIAGGLAMLLEIYDFPPYEGLLDAHAL 382
V + +Q ++W+ W W W +VV +AM LE+ DFPP G +DAH+L
Sbjct: 254 VAVGAIQNLLWSWYSWTRYREQKKGWAAWPGIVVAWVLVAMSLELLDFPPLWGSVDAHSL 313
Query: 381 WHATTIPLTYIWWSFIRDDAE 319
WHA TI T IW++F+ DA+
Sbjct: 314 WHAGTIVPTIIWYNFLVKDAQ 334
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 493,110,542
Number of Sequences: 1393205
Number of extensions: 10713120
Number of successful extensions: 32338
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 31477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32318
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)