Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001532A_C02 KMC001532A_c02
(564 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
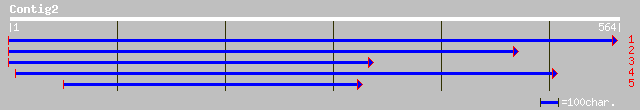
ref|NP_189573.1| unknown protein; protein id: At3g29270.1 [Arabi... 91 8e-18
dbj|BAB03123.1| RING zinc finger protein-like [Arabidopsis thali... 91 8e-18
ref|NP_177577.1| putative RING zinc finger protein; protein id: ... 75 6e-13
gb|AAM64512.1| putative RING zinc finger protein [Arabidopsis th... 75 6e-13
gb|AAM65966.1| unknown [Arabidopsis thaliana] 71 1e-11
>ref|NP_189573.1| unknown protein; protein id: At3g29270.1 [Arabidopsis thaliana]
gi|27754358|gb|AAO22628.1| unknown protein [Arabidopsis
thaliana] gi|28394061|gb|AAO42438.1| unknown protein
[Arabidopsis thaliana]
Length = 263
Score = 91.3 bits (225), Expect = 8e-18
Identities = 43/53 (81%), Positives = 49/53 (92%)
Frame = -1
Query: 564 VIIFLLIILYAIPASAVILALYILITILFALPSFLILYFSYPSLDWLVREIIT 406
V+IFLLIILYAIP SA ILA+YIL+T+L ALPSFLILYF+YP LDWLVREI+T
Sbjct: 211 VVIFLLIILYAIPTSAAILAMYILVTLLLALPSFLILYFAYPCLDWLVREIVT 263
>dbj|BAB03123.1| RING zinc finger protein-like [Arabidopsis thaliana]
Length = 255
Score = 91.3 bits (225), Expect = 8e-18
Identities = 43/53 (81%), Positives = 49/53 (92%)
Frame = -1
Query: 564 VIIFLLIILYAIPASAVILALYILITILFALPSFLILYFSYPSLDWLVREIIT 406
V+IFLLIILYAIP SA ILA+YIL+T+L ALPSFLILYF+YP LDWLVREI+T
Sbjct: 203 VVIFLLIILYAIPTSAAILAMYILVTLLLALPSFLILYFAYPCLDWLVREIVT 255
>ref|NP_177577.1| putative RING zinc finger protein; protein id: At1g74370.1,
supported by cDNA: 29883. [Arabidopsis thaliana]
gi|25373335|pir||D96772 probable RING zinc finger
protein F1M20.5 [imported] - Arabidopsis thaliana
gi|12324808|gb|AAG52370.1|AC011765_22 putative RING zinc
finger protein; 15305-14520 [Arabidopsis thaliana]
Length = 261
Score = 75.1 bits (183), Expect = 6e-13
Identities = 32/53 (60%), Positives = 43/53 (80%)
Frame = -1
Query: 564 VIIFLLIILYAIPASAVILALYILITILFALPSFLILYFSYPSLDWLVREIIT 406
V+IFLL+ LYAIP SA +L +Y +T A+PSFL+LYF++PSL+WL+REI T
Sbjct: 209 VVIFLLMALYAIPVSAAVLGVYFFVTFALAVPSFLVLYFAFPSLNWLIREIAT 261
>gb|AAM64512.1| putative RING zinc finger protein [Arabidopsis thaliana]
Length = 261
Score = 75.1 bits (183), Expect = 6e-13
Identities = 32/53 (60%), Positives = 43/53 (80%)
Frame = -1
Query: 564 VIIFLLIILYAIPASAVILALYILITILFALPSFLILYFSYPSLDWLVREIIT 406
V+IFLL+ LYAIP SA +L +Y +T A+PSFL+LYF++PSL+WL+REI T
Sbjct: 209 VVIFLLMALYAIPVSAAVLGVYFFVTFALAVPSFLVLYFAFPSLNWLIREIAT 261
>gb|AAM65966.1| unknown [Arabidopsis thaliana]
Length = 270
Score = 70.9 bits (172), Expect = 1e-11
Identities = 34/51 (66%), Positives = 43/51 (83%)
Frame = -1
Query: 564 VIIFLLIILYAIPASAVILALYILITILFALPSFLILYFSYPSLDWLVREI 412
VIIFLLI+ +AIP S +ILALY L+TILFA+P+ L+LYF+YP L+ LV EI
Sbjct: 217 VIIFLLIVFFAIPGSLIILALYFLLTILFAVPAGLVLYFAYPILERLVNEI 267
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 465,583,290
Number of Sequences: 1393205
Number of extensions: 9746860
Number of successful extensions: 24117
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 23571
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24107
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)