Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001494A_C01 KMC001494A_c01
(642 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
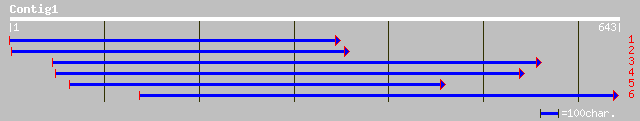
ref|NP_564658.1| expressed protein; protein id: At1g54390.1, sup... 146 2e-34
pir||G96585 hypothetical protein F20D21.22 [imported] - Arabidop... 146 2e-34
ref|NP_566742.1| PHD-finger protein, putative; protein id: At3g2... 99 4e-20
gb|AAO42002.1| putative PHD-finger protein [Arabidopsis thaliana] 99 4e-20
gb|EAA14095.1| agCP8300 [Anopheles gambiae str. PEST] 99 5e-20
>ref|NP_564658.1| expressed protein; protein id: At1g54390.1, supported by cDNA:
157730. [Arabidopsis thaliana]
gi|21553698|gb|AAM62791.1| putative growth inhibitory
protein [Arabidopsis thaliana]
Length = 181
Score = 146 bits (369), Expect = 2e-34
Identities = 63/74 (85%), Positives = 70/74 (94%)
Frame = -1
Query: 642 HKKDYVIPMDVDQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYACVGLTQETRF 463
++KD ++P++ +QPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHY CVGLT ETRF
Sbjct: 108 NRKD-LMPIE-EQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYTCVGLTPETRF 165
Query: 462 KGKWYCPTCRLLPQ 421
KGKWYCPTCRLLPQ
Sbjct: 166 KGKWYCPTCRLLPQ 179
>pir||G96585 hypothetical protein F20D21.22 [imported] - Arabidopsis thaliana
gi|4585982|gb|AAD25618.1|AC005287_20 Unknown protein
[Arabidopsis thaliana]
Length = 232
Score = 146 bits (369), Expect = 2e-34
Identities = 63/74 (85%), Positives = 70/74 (94%)
Frame = -1
Query: 642 HKKDYVIPMDVDQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYACVGLTQETRF 463
++KD ++P++ +QPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHY CVGLT ETRF
Sbjct: 159 NRKD-LMPIE-EQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYTCVGLTPETRF 216
Query: 462 KGKWYCPTCRLLPQ 421
KGKWYCPTCRLLPQ
Sbjct: 217 KGKWYCPTCRLLPQ 230
>ref|NP_566742.1| PHD-finger protein, putative; protein id: At3g24010.1, supported by
cDNA: 95660. [Arabidopsis thaliana]
gi|9294670|dbj|BAB03019.1| gene_id:F14O13.20~unknown
protein [Arabidopsis thaliana]
gi|21618065|gb|AAM67115.1| PHD-finger protein, putative
[Arabidopsis thaliana]
Length = 234
Score = 99.4 bits (246), Expect = 4e-20
Identities = 40/61 (65%), Positives = 50/61 (81%)
Frame = -1
Query: 618 MDVDQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYACVGLTQETRFKGKWYCPT 439
MD+D P+DPNEPTYC+C+QVSFG+M+ACDN C+ EWFH+ CVGL ++ KGKWYCP
Sbjct: 167 MDLDLPVDPNEPTYCICNQVSFGEMVACDNNACK-IEWFHFGCVGLKEQP--KGKWYCPE 223
Query: 438 C 436
C
Sbjct: 224 C 224
>gb|AAO42002.1| putative PHD-finger protein [Arabidopsis thaliana]
Length = 160
Score = 99.4 bits (246), Expect = 4e-20
Identities = 40/61 (65%), Positives = 50/61 (81%)
Frame = -1
Query: 618 MDVDQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYACVGLTQETRFKGKWYCPT 439
MD+D P+DPNEPTYC+C+QVSFG+M+ACDN C+ EWFH+ CVGL ++ KGKWYCP
Sbjct: 93 MDLDLPVDPNEPTYCICNQVSFGEMVACDNNACK-IEWFHFGCVGLKEQP--KGKWYCPE 149
Query: 438 C 436
C
Sbjct: 150 C 150
>gb|EAA14095.1| agCP8300 [Anopheles gambiae str. PEST]
Length = 295
Score = 99.0 bits (245), Expect = 5e-20
Identities = 43/63 (68%), Positives = 51/63 (80%), Gaps = 1/63 (1%)
Frame = -1
Query: 621 PMDV-DQPIDPNEPTYCVCHQVSFGDMIACDNENCQGGEWFHYACVGLTQETRFKGKWYC 445
P DV D P+DPNEPTYC+CHQVS+G+MI CDN +C EWFH+ACVGLT T+ KGKW+C
Sbjct: 229 PSDVLDMPVDPNEPTYCLCHQVSYGEMIGCDNPDCP-IEWFHFACVGLT--TKPKGKWFC 285
Query: 444 PTC 436
P C
Sbjct: 286 PKC 288
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,645,115
Number of Sequences: 1393205
Number of extensions: 12607832
Number of successful extensions: 39258
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 37129
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39072
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)